inst/examples/misc_examples/mietchen.md
In cboettig/wrightscape: wrightscape
mietchen.R
load some useful libraries and the phylogeny
require(auteur)
data(primates)
load in Daniel's data
dat <- read.csv("../../../data/Primate_brain_comparisons.csv")
Concatenate Genus and species names
SpeciesNames <- sapply(1:length(dat[[1]]),
function(i)
paste(dat[[1]][i], "_", dat[[2]][i], sep=""))
seems these two genus names differ in spelling in the data and tree,
let's just fix them manually
SpeciesNames <- gsub("Saguinas", "Saguinus", SpeciesNames)
SpeciesNames <- gsub("Presbytus", "Presbytis", SpeciesNames)
Name the data rows as "Genus_species", matching the tree tip label convention
rownames(dat) <- SpeciesNames
get just the quantitative trait data
dat <- dat[3:7]
drop all tips that don't have data, or data that doesn't have tips in tree
primate_data <- treedata(primates$phy, dat)
Dropped tips from the tree because there were no matching names in the data:
[1] "Allenopithecus_nigroviridis" "Allocebus_trichotis"
[3] "Alouatta_belzebul" "Alouatta_caraya"
[5] "Alouatta_coibensis" "Alouatta_fusca"
[7] "Alouatta_pigra" "Alouatta_sara"
[9] "Aotus_azarai" "Aotus_brumbacki"
[11] "Aotus_hershkovitzi" "Aotus_infulatus"
[13] "Aotus_lemurinus" "Aotus_miconax"
[15] "Aotus_nancymaae" "Aotus_nigriceps"
[17] "Aotus_vociferans" "Arctocebus_aureus"
[19] "Arctocebus_calabarensis" "Ateles_chamek"
[21] "Ateles_fusciceps" "Ateles_marginatus"
[23] "Ateles_paniscus" "Avahi_laniger"
[25] "Brachyteles_arachnoides" "Cacajao_calvus"
[27] "Cacajao_melanocephalus" "Callicebus_brunneus"
[29] "Callicebus_caligatus" "Callicebus_cinerascens"
[31] "Callicebus_donacophilus" "Callicebus_dubius"
[33] "Callicebus_hoffmannsi" "Callicebus_modestus"
[35] "Callicebus_oenanthe" "Callicebus_olallae"
[37] "Callicebus_personatus" "Callicebus_torquatus"
[39] "Callimico_goeldii" "Callithrix_argentata"
[41] "Callithrix_aurita" "Callithrix_flaviceps"
[43] "Callithrix_humeralifera" "Callithrix_kuhlii"
[45] "Callithrix_penicillata" "Callithrix_pygmaea"
[47] "Cebus_olivaceus" "Cercocebus_agilis"
[49] "Cercopithecus_campbelli" "Cercopithecus_cephus"
[51] "Cercopithecus_diana" "Cercopithecus_dryas"
[53] "Cercopithecus_erythrogaster" "Cercopithecus_erythrotis"
[55] "Cercopithecus_neglectus" "Cercopithecus_nictitans"
[57] "Cercopithecus_petaurista" "Cercopithecus_pogonias"
[59] "Cercopithecus_preussi" "Cercopithecus_sclateri"
[61] "Cercopithecus_solatus" "Cercopithecus_wolfi"
[63] "Cheirogaleus_major" "Cheirogaleus_medius"
[65] "Chiropotes_albinasus" "Chiropotes_satanas"
[67] "Chlorocebus_aethiops" "Colobus_angolensis"
[69] "Colobus_guereza" "Colobus_polykomos"
[71] "Colobus_satanas" "Daubentonia_madagascariensis"
[73] "Eulemur_coronatus" "Eulemur_fulvus"
[75] "Eulemur_macaco" "Eulemur_mongoz"
[77] "Eulemur_rubriventer" "Euoticus_elegantulus"
[79] "Euoticus_pallidus" "Galago_alleni"
[81] "Galago_gallarum" "Galago_matschiei"
[83] "Galago_moholi" "Galago_senegalensis"
[85] "Galagoides_demidoff" "Galagoides_zanzibaricus"
[87] "Hapalemur_aureus" "Hapalemur_griseus"
[89] "Hapalemur_simus" "Hylobates_concolor"
[91] "Hylobates_gabriellae" "Hylobates_hoolock"
[93] "Hylobates_klossii" "Hylobates_leucogenys"
[95] "Hylobates_muelleri" "Hylobates_syndactylus"
[97] "Indri_indri" "Lagothrix_flavicauda"
[99] "Lagothrix_lagotricha" "Lemur_catta"
[101] "Leontopithecus_caissara" "Leontopithecus_chrysomela"
[103] "Leontopithecus_chrysopygus" "Leontopithecus_rosalia"
[105] "Lepilemur_dorsalis" "Lepilemur_edwardsi"
[107] "Lepilemur_leucopus" "Lepilemur_microdon"
[109] "Lepilemur_mustelinus" "Lepilemur_ruficaudatus"
[111] "Lepilemur_septentrionalis" "Lophocebus_albigena"
[113] "Loris_tardigradus" "Macaca_assamensis"
[115] "Macaca_cyclopis" "Macaca_fuscata"
[117] "Macaca_maura" "Macaca_nigra"
[119] "Macaca_ochreata" "Macaca_radiata"
[121] "Macaca_silenus" "Macaca_sylvanus"
[123] "Macaca_thibetana" "Macaca_tonkeana"
[125] "Mandrillus_leucophaeus" "Microcebus_coquereli"
[127] "Microcebus_murinus" "Microcebus_rufus"
[129] "Nasalis_concolor" "Nycticebus_coucang"
[131] "Nycticebus_pygmaeus" "Otolemur_crassicaudatus"
[133] "Otolemur_garnettii" "Perodicticus_potto"
[135] "Phaner_furcifer" "Pithecia_aequatorialis"
[137] "Pithecia_albicans" "Pithecia_irrorata"
[139] "Pithecia_pithecia" "Presbytis_comata"
[141] "Presbytis_femoralis" "Presbytis_frontata"
[143] "Presbytis_hosei" "Presbytis_melalophos"
[145] "Presbytis_potenziani" "Presbytis_rubicunda"
[147] "Presbytis_thomasi" "Procolobus_badius"
[149] "Procolobus_pennantii" "Procolobus_preussi"
[151] "Procolobus_rufomitratus" "Procolobus_verus"
[153] "Propithecus_diadema" "Propithecus_tattersalli"
[155] "Propithecus_verreauxi" "Pygathrix_avunculus"
[157] "Pygathrix_bieti" "Pygathrix_brelichi"
[159] "Pygathrix_roxellana" "Saguinus_bicolor"
[161] "Saguinus_fuscicollis" "Saguinus_geoffroyi"
[163] "Saguinus_imperator" "Saguinus_inustus"
[165] "Saguinus_labiatus" "Saguinus_leucopus"
[167] "Saguinus_mystax" "Saguinus_nigricollis"
[169] "Saguinus_tripartitus" "Saimiri_boliviensis"
[171] "Saimiri_ustus" "Saimiri_vanzolinii"
[173] "Semnopithecus_entellus" "Tarsius_bancanus"
[175] "Tarsius_dianae" "Tarsius_pumilus"
[177] "Tarsius_spectrum" "Tarsius_syrichta"
[179] "Theropithecus_gelada" "Trachypithecus_auratus"
[181] "Trachypithecus_cristatus" "Trachypithecus_francoisi"
[183] "Trachypithecus_geei" "Trachypithecus_johnii"
[185] "Trachypithecus_obscurus" "Trachypithecus_phayrei"
[187] "Trachypithecus_pileatus" "Trachypithecus_vetulus"
[189] "Varecia_variegata"
Dropped rows from the data because there were no matching tips in the tree:
[1] "Alouatta_villosa" "Callimico_goeldi"
[3] "Cebuella_pygmaea" "Cercocebus_albigena"
[5] "Cercopithecus_aethiops" "Cercopithecus_pygerythrus"
[7] "Colobus_badius" "Lagothrix_lagothrica"
[9] "Papio_anubis" "Papio_cynocephalus"
[11] "Papio_papio" "Papio_ursinus"
[13] "Presbytis_cristatus" "Presbytis_entellus"
[15] "Presbytis_obscurus" "Saguinus_tamarin"
[17] "Symphalangus_syndactylus"
dat <- primate_data$data
phy <- primate_data$phy
Classic independent contrasts for a phylogenetic correction to the
estimate of correlations in brain weight with body size
x <- pic(log(dat[,"Body_weight"]), phy)
y <- pic(log(dat[,"Brain_weight"]), phy)
summary(lm(y~x-1))
Call:
lm(formula = y ~ x - 1)
Residuals:
Min 1Q Median 3Q Max
-0.2599 -0.0330 0.0099 0.0413 0.3711
Coefficients:
Estimate Std. Error t value Pr(>|t|)
x 0.6408 0.0562 11.4 2e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.0983 on 42 degrees of freedom
Multiple R-squared: 0.756, Adjusted R-squared: 0.75
F-statistic: 130 on 1 and 42 DF, p-value: 1.99e-14
get all ancestral states. Note that Ancestral state estimates
are highly uncertain and generally distrusted, see Schluter et al 1997.
ancestral_states <- lapply(1:dim(dat)[2], function(i) ace(dat[,i], phy))
get the BM diversification rates for each trait
diversification_rates <- fitContinuous(phy, dat)
Fitting BM model:
Estimate a shift in diversification rate using AUTEUR
run two short reversible-jump Markov chains
(create some random strings for temporary file names)
r=paste(sample(letters,9,replace=TRUE),collapse="")
run four short MCMC chains to search for a change point in brain weight
require(snowfall)
sfInit(parallel=TRUE, cpu=4)
R Version: R version 2.15.0 (2012-03-30)
sfLibrary(auteur)
Library auteur loaded.
sfExportAll()
out <- sfLapply(1:4,
function(x) rjmcmc.bm(phy=phy, dat=dat[,"log_brain.weight"],
ngen=100000, sample.freq=10, prob.mergesplit=0.1, simplestart=TRUE,
prop.width=1, fileBase=paste(r,x,sep=".")))
collect directories
dirs=dir("./",pattern=paste("BM",r,sep="."))
pool.rjmcmcsamples(base.dirs=dirs, lab=r)
view contents of .rda
load(paste(paste(r,"combined.rjmcmc",sep="."),
paste(r,"posteriorsamples.rda",sep="."),sep="/"))
print(head(posteriorsamples$rates))
46 47 48 38 49 6 7 50 51
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
1 2 52 4 5 53 54 55 11
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
56 12 10 57 44 43 58 3 59
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 20.58079 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
60 8 9 61 42 41 62 63 64
1 0.06713 14.8483 0.06713 0.06713 0.06713 24.2739 0.06713 0.06713 0.06713
2 1.00000 1.0000 1.00000 1.00000 1.00000 1.0000 1.00000 1.00000 1.00000
3 0.22906 0.2291 0.22906 0.22906 0.22906 0.2291 0.22906 0.22906 0.22906
4 0.14813 0.1481 0.14813 0.14813 0.14813 0.1481 0.14813 0.14813 0.14813
5 0.03146 14.8483 0.03146 0.03146 0.03146 24.2739 0.03146 0.03146 0.03146
6 0.37253 0.3725 0.37253 0.37253 0.37253 0.3725 0.37253 0.37253 0.37253
39 65 21 66 22 67 36 35 68
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
69 25 70 24 23 26 71 72 40
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
34 73 74 75 76 32 77 14 13
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 2.99136 0.03146 0.03146 0.03146
6 0.37253 0.37253 3.54680 3.54680 3.54680 3.54680 3.54680 3.54680 3.54680
37 78 30 79 80 27 31 81 28
1 0.06713 0.06713 0.06713 0.06713 0.06713 12.3205 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.0000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 6.79323 6.79323 6.7932 6.79323 6.79323 6.79323
4 0.14813 0.14813 73.40918 0.14813 0.14813 0.1481 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 12.3205 0.03146 0.03146 0.03146
6 3.54680 3.54680 3.54680 3.54680 3.54680 3.5468 3.54680 3.54680 3.54680
29 82 33 83 20 84 17 85 16
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 6.79323 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 3.54680 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
86 19 87 15 18
1 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253
print(head(posteriorsamples$rate.shifts))
NULL
plot Markov sampled rates
shifts.plot(phy=phy, base.dir=paste(r,"combined.rjmcmc",sep="."), burnin=0.5, legend=TRUE, edge.width=4, x.lim = c(0,60))
READING estimates...
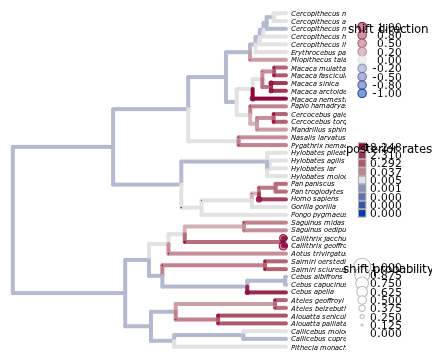
$res
branch shift.direction shift.probability
1 46 0.0000 0.00000
2 47 0.0000 0.00000
3 48 0.0000 0.00000
4 38 0.0000 0.00000
5 49 0.0000 0.00000
6 6 0.0000 0.00000
7 7 0.0000 0.00000
8 50 0.0000 0.00000
9 51 0.9318 0.01905
10 1 0.9990 0.10000
11 2 1.0000 0.14160
12 52 0.0000 0.00000
13 4 0.9958 0.04815
14 5 1.0000 0.08125
15 53 0.0000 0.00000
16 54 0.0000 0.00000
17 55 0.0000 0.00000
18 11 1.0000 0.25095
19 56 0.0000 0.00000
20 12 0.0000 0.00000
21 10 0.0000 0.00000
22 57 0.9895 0.02870
23 44 0.9847 0.09830
24 43 0.9980 0.09920
25 58 0.0000 0.00000
26 3 0.9784 0.02780
27 59 0.0000 0.00000
28 60 0.9780 0.10005
29 8 0.9716 0.48660
30 9 1.0000 0.42810
31 61 0.0000 0.00000
32 42 0.9423 0.02080
33 41 0.9653 0.03170
34 62 0.0000 0.00000
35 63 0.0000 0.00000
36 64 0.9923 0.03905
37 39 0.0000 0.00000
38 65 1.0000 0.01215
39 21 0.0000 0.00000
40 66 1.0000 0.02290
41 22 0.9988 0.32165
42 67 0.9846 0.03900
43 36 0.0000 0.00000
44 35 0.0000 0.00000
45 68 0.0000 0.00000
46 69 0.0000 0.00000
47 25 0.0000 0.00000
48 70 0.0000 0.00000
49 24 0.0000 0.00000
50 23 0.0000 0.00000
51 26 0.0000 0.00000
52 71 0.0000 0.00000
53 72 0.0000 0.00000
54 40 0.9855 0.03440
55 34 1.0000 0.03155
56 73 0.0000 0.00000
57 74 0.0000 0.00000
58 75 0.0000 0.00000
59 76 0.0000 0.00000
60 32 0.8537 0.01025
61 77 0.9472 0.01705
62 14 0.9994 0.15475
63 13 0.9977 0.13260
64 37 0.8816 0.02955
65 78 0.0000 0.00000
66 30 0.9990 0.28895
67 79 0.0000 0.00000
68 80 0.8462 0.01495
69 27 1.0000 0.23400
70 31 0.9980 0.25450
71 81 0.9052 0.01160
72 28 0.9974 0.11755
73 29 0.9800 0.09005
74 82 0.0000 0.00000
75 33 0.9836 0.01830
76 83 0.0000 0.00000
77 20 0.0000 0.00000
78 84 0.0000 0.00000
79 17 0.0000 0.00000
80 85 0.0000 0.00000
81 16 0.0000 0.00000
82 86 0.0000 0.00000
83 19 0.0000 0.00000
84 87 0.0000 0.00000
85 15 0.0000 0.00000
86 18 0.0000 0.00000
$desc
$desc$descendants
$desc$descendants$`60`
[1] "Callithrix_geoffroyi" "Callithrix_jacchus"
$desc$descendants$`64`
[1] "Pongo_pygmaeus" "Gorilla_gorilla" "Homo_sapiens" "Pan_troglodytes"
[5] "Pan_paniscus"
$desc$descendants$`67`
[1] "Pan_troglodytes" "Pan_paniscus"
$desc$descendants$`57`
[1] "Saimiri_sciureus" "Saimiri_oerstedii"
$desc$descendants$`66`
[1] "Homo_sapiens" "Pan_troglodytes" "Pan_paniscus"
$desc$descendants$`51`
[1] "Alouatta_palliata" "Alouatta_seniculus"
$desc$descendants$`77`
[1] "Cercocebus_torquatus" "Cercocebus_galeritus"
$desc$descendants$`80`
[1] "Macaca_arctoides" "Macaca_sinica"
$desc$descendants$`65`
[1] "Gorilla_gorilla" "Homo_sapiens" "Pan_troglodytes" "Pan_paniscus"
$desc$descendants$`81`
[1] "Macaca_fascicularis" "Macaca_mulatta"
$desc$branch.shift.probability
8 9 22 30 31 11 27 14 2
0.48660 0.42810 0.32165 0.28895 0.25450 0.25095 0.23400 0.15475 0.14160
13 28 60 1 43 44 29 5 4
0.13260 0.11755 0.10005 0.10000 0.09920 0.09830 0.09005 0.08125 0.04815
64 67 40 41 34 37 57 3 66
0.03905 0.03900 0.03440 0.03170 0.03155 0.02955 0.02870 0.02780 0.02290
42 51 33 77 80 65 81 32
0.02080 0.01905 0.01830 0.01705 0.01495 0.01215 0.01160 0.01025
cboettig/wrightscape documentation built on May 13, 2019, 2:12 p.m.
mietchen.R
load some useful libraries and the phylogeny
require(auteur)
data(primates)
load in Daniel's data
dat <- read.csv("../../../data/Primate_brain_comparisons.csv")
Concatenate Genus and species names
SpeciesNames <- sapply(1:length(dat[[1]]),
function(i)
paste(dat[[1]][i], "_", dat[[2]][i], sep=""))
seems these two genus names differ in spelling in the data and tree, let's just fix them manually
SpeciesNames <- gsub("Saguinas", "Saguinus", SpeciesNames)
SpeciesNames <- gsub("Presbytus", "Presbytis", SpeciesNames)
Name the data rows as "Genus_species", matching the tree tip label convention
rownames(dat) <- SpeciesNames
get just the quantitative trait data
dat <- dat[3:7]
drop all tips that don't have data, or data that doesn't have tips in tree
primate_data <- treedata(primates$phy, dat)
Dropped tips from the tree because there were no matching names in the data:
[1] "Allenopithecus_nigroviridis" "Allocebus_trichotis"
[3] "Alouatta_belzebul" "Alouatta_caraya"
[5] "Alouatta_coibensis" "Alouatta_fusca"
[7] "Alouatta_pigra" "Alouatta_sara"
[9] "Aotus_azarai" "Aotus_brumbacki"
[11] "Aotus_hershkovitzi" "Aotus_infulatus"
[13] "Aotus_lemurinus" "Aotus_miconax"
[15] "Aotus_nancymaae" "Aotus_nigriceps"
[17] "Aotus_vociferans" "Arctocebus_aureus"
[19] "Arctocebus_calabarensis" "Ateles_chamek"
[21] "Ateles_fusciceps" "Ateles_marginatus"
[23] "Ateles_paniscus" "Avahi_laniger"
[25] "Brachyteles_arachnoides" "Cacajao_calvus"
[27] "Cacajao_melanocephalus" "Callicebus_brunneus"
[29] "Callicebus_caligatus" "Callicebus_cinerascens"
[31] "Callicebus_donacophilus" "Callicebus_dubius"
[33] "Callicebus_hoffmannsi" "Callicebus_modestus"
[35] "Callicebus_oenanthe" "Callicebus_olallae"
[37] "Callicebus_personatus" "Callicebus_torquatus"
[39] "Callimico_goeldii" "Callithrix_argentata"
[41] "Callithrix_aurita" "Callithrix_flaviceps"
[43] "Callithrix_humeralifera" "Callithrix_kuhlii"
[45] "Callithrix_penicillata" "Callithrix_pygmaea"
[47] "Cebus_olivaceus" "Cercocebus_agilis"
[49] "Cercopithecus_campbelli" "Cercopithecus_cephus"
[51] "Cercopithecus_diana" "Cercopithecus_dryas"
[53] "Cercopithecus_erythrogaster" "Cercopithecus_erythrotis"
[55] "Cercopithecus_neglectus" "Cercopithecus_nictitans"
[57] "Cercopithecus_petaurista" "Cercopithecus_pogonias"
[59] "Cercopithecus_preussi" "Cercopithecus_sclateri"
[61] "Cercopithecus_solatus" "Cercopithecus_wolfi"
[63] "Cheirogaleus_major" "Cheirogaleus_medius"
[65] "Chiropotes_albinasus" "Chiropotes_satanas"
[67] "Chlorocebus_aethiops" "Colobus_angolensis"
[69] "Colobus_guereza" "Colobus_polykomos"
[71] "Colobus_satanas" "Daubentonia_madagascariensis"
[73] "Eulemur_coronatus" "Eulemur_fulvus"
[75] "Eulemur_macaco" "Eulemur_mongoz"
[77] "Eulemur_rubriventer" "Euoticus_elegantulus"
[79] "Euoticus_pallidus" "Galago_alleni"
[81] "Galago_gallarum" "Galago_matschiei"
[83] "Galago_moholi" "Galago_senegalensis"
[85] "Galagoides_demidoff" "Galagoides_zanzibaricus"
[87] "Hapalemur_aureus" "Hapalemur_griseus"
[89] "Hapalemur_simus" "Hylobates_concolor"
[91] "Hylobates_gabriellae" "Hylobates_hoolock"
[93] "Hylobates_klossii" "Hylobates_leucogenys"
[95] "Hylobates_muelleri" "Hylobates_syndactylus"
[97] "Indri_indri" "Lagothrix_flavicauda"
[99] "Lagothrix_lagotricha" "Lemur_catta"
[101] "Leontopithecus_caissara" "Leontopithecus_chrysomela"
[103] "Leontopithecus_chrysopygus" "Leontopithecus_rosalia"
[105] "Lepilemur_dorsalis" "Lepilemur_edwardsi"
[107] "Lepilemur_leucopus" "Lepilemur_microdon"
[109] "Lepilemur_mustelinus" "Lepilemur_ruficaudatus"
[111] "Lepilemur_septentrionalis" "Lophocebus_albigena"
[113] "Loris_tardigradus" "Macaca_assamensis"
[115] "Macaca_cyclopis" "Macaca_fuscata"
[117] "Macaca_maura" "Macaca_nigra"
[119] "Macaca_ochreata" "Macaca_radiata"
[121] "Macaca_silenus" "Macaca_sylvanus"
[123] "Macaca_thibetana" "Macaca_tonkeana"
[125] "Mandrillus_leucophaeus" "Microcebus_coquereli"
[127] "Microcebus_murinus" "Microcebus_rufus"
[129] "Nasalis_concolor" "Nycticebus_coucang"
[131] "Nycticebus_pygmaeus" "Otolemur_crassicaudatus"
[133] "Otolemur_garnettii" "Perodicticus_potto"
[135] "Phaner_furcifer" "Pithecia_aequatorialis"
[137] "Pithecia_albicans" "Pithecia_irrorata"
[139] "Pithecia_pithecia" "Presbytis_comata"
[141] "Presbytis_femoralis" "Presbytis_frontata"
[143] "Presbytis_hosei" "Presbytis_melalophos"
[145] "Presbytis_potenziani" "Presbytis_rubicunda"
[147] "Presbytis_thomasi" "Procolobus_badius"
[149] "Procolobus_pennantii" "Procolobus_preussi"
[151] "Procolobus_rufomitratus" "Procolobus_verus"
[153] "Propithecus_diadema" "Propithecus_tattersalli"
[155] "Propithecus_verreauxi" "Pygathrix_avunculus"
[157] "Pygathrix_bieti" "Pygathrix_brelichi"
[159] "Pygathrix_roxellana" "Saguinus_bicolor"
[161] "Saguinus_fuscicollis" "Saguinus_geoffroyi"
[163] "Saguinus_imperator" "Saguinus_inustus"
[165] "Saguinus_labiatus" "Saguinus_leucopus"
[167] "Saguinus_mystax" "Saguinus_nigricollis"
[169] "Saguinus_tripartitus" "Saimiri_boliviensis"
[171] "Saimiri_ustus" "Saimiri_vanzolinii"
[173] "Semnopithecus_entellus" "Tarsius_bancanus"
[175] "Tarsius_dianae" "Tarsius_pumilus"
[177] "Tarsius_spectrum" "Tarsius_syrichta"
[179] "Theropithecus_gelada" "Trachypithecus_auratus"
[181] "Trachypithecus_cristatus" "Trachypithecus_francoisi"
[183] "Trachypithecus_geei" "Trachypithecus_johnii"
[185] "Trachypithecus_obscurus" "Trachypithecus_phayrei"
[187] "Trachypithecus_pileatus" "Trachypithecus_vetulus"
[189] "Varecia_variegata"
Dropped rows from the data because there were no matching tips in the tree:
[1] "Alouatta_villosa" "Callimico_goeldi"
[3] "Cebuella_pygmaea" "Cercocebus_albigena"
[5] "Cercopithecus_aethiops" "Cercopithecus_pygerythrus"
[7] "Colobus_badius" "Lagothrix_lagothrica"
[9] "Papio_anubis" "Papio_cynocephalus"
[11] "Papio_papio" "Papio_ursinus"
[13] "Presbytis_cristatus" "Presbytis_entellus"
[15] "Presbytis_obscurus" "Saguinus_tamarin"
[17] "Symphalangus_syndactylus"
dat <- primate_data$data
phy <- primate_data$phy
Classic independent contrasts for a phylogenetic correction to the estimate of correlations in brain weight with body size
x <- pic(log(dat[,"Body_weight"]), phy)
y <- pic(log(dat[,"Brain_weight"]), phy)
summary(lm(y~x-1))
Call:
lm(formula = y ~ x - 1)
Residuals:
Min 1Q Median 3Q Max
-0.2599 -0.0330 0.0099 0.0413 0.3711
Coefficients:
Estimate Std. Error t value Pr(>|t|)
x 0.6408 0.0562 11.4 2e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.0983 on 42 degrees of freedom
Multiple R-squared: 0.756, Adjusted R-squared: 0.75
F-statistic: 130 on 1 and 42 DF, p-value: 1.99e-14
get all ancestral states. Note that Ancestral state estimates are highly uncertain and generally distrusted, see Schluter et al 1997.
ancestral_states <- lapply(1:dim(dat)[2], function(i) ace(dat[,i], phy))
get the BM diversification rates for each trait
diversification_rates <- fitContinuous(phy, dat)
Fitting BM model:
Estimate a shift in diversification rate using AUTEUR
run two short reversible-jump Markov chains (create some random strings for temporary file names)
r=paste(sample(letters,9,replace=TRUE),collapse="")
run four short MCMC chains to search for a change point in brain weight
require(snowfall)
sfInit(parallel=TRUE, cpu=4)
R Version: R version 2.15.0 (2012-03-30)
sfLibrary(auteur)
Library auteur loaded.
sfExportAll()
out <- sfLapply(1:4,
function(x) rjmcmc.bm(phy=phy, dat=dat[,"log_brain.weight"],
ngen=100000, sample.freq=10, prob.mergesplit=0.1, simplestart=TRUE,
prop.width=1, fileBase=paste(r,x,sep=".")))
collect directories
dirs=dir("./",pattern=paste("BM",r,sep="."))
pool.rjmcmcsamples(base.dirs=dirs, lab=r)
view contents of .rda
load(paste(paste(r,"combined.rjmcmc",sep="."),
paste(r,"posteriorsamples.rda",sep="."),sep="/"))
print(head(posteriorsamples$rates))
46 47 48 38 49 6 7 50 51
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
1 2 52 4 5 53 54 55 11
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
56 12 10 57 44 43 58 3 59
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 20.58079 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
60 8 9 61 42 41 62 63 64
1 0.06713 14.8483 0.06713 0.06713 0.06713 24.2739 0.06713 0.06713 0.06713
2 1.00000 1.0000 1.00000 1.00000 1.00000 1.0000 1.00000 1.00000 1.00000
3 0.22906 0.2291 0.22906 0.22906 0.22906 0.2291 0.22906 0.22906 0.22906
4 0.14813 0.1481 0.14813 0.14813 0.14813 0.1481 0.14813 0.14813 0.14813
5 0.03146 14.8483 0.03146 0.03146 0.03146 24.2739 0.03146 0.03146 0.03146
6 0.37253 0.3725 0.37253 0.37253 0.37253 0.3725 0.37253 0.37253 0.37253
39 65 21 66 22 67 36 35 68
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
69 25 70 24 23 26 71 72 40
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
34 73 74 75 76 32 77 14 13
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 2.99136 0.03146 0.03146 0.03146
6 0.37253 0.37253 3.54680 3.54680 3.54680 3.54680 3.54680 3.54680 3.54680
37 78 30 79 80 27 31 81 28
1 0.06713 0.06713 0.06713 0.06713 0.06713 12.3205 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.0000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 6.79323 6.79323 6.7932 6.79323 6.79323 6.79323
4 0.14813 0.14813 73.40918 0.14813 0.14813 0.1481 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 12.3205 0.03146 0.03146 0.03146
6 3.54680 3.54680 3.54680 3.54680 3.54680 3.5468 3.54680 3.54680 3.54680
29 82 33 83 20 84 17 85 16
1 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
3 6.79323 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146 0.03146
6 3.54680 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253 0.37253
86 19 87 15 18
1 0.06713 0.06713 0.06713 0.06713 0.06713
2 1.00000 1.00000 1.00000 1.00000 1.00000
3 0.22906 0.22906 0.22906 0.22906 0.22906
4 0.14813 0.14813 0.14813 0.14813 0.14813
5 0.03146 0.03146 0.03146 0.03146 0.03146
6 0.37253 0.37253 0.37253 0.37253 0.37253
print(head(posteriorsamples$rate.shifts))
NULL
plot Markov sampled rates
shifts.plot(phy=phy, base.dir=paste(r,"combined.rjmcmc",sep="."), burnin=0.5, legend=TRUE, edge.width=4, x.lim = c(0,60))
READING estimates...

$res
branch shift.direction shift.probability
1 46 0.0000 0.00000
2 47 0.0000 0.00000
3 48 0.0000 0.00000
4 38 0.0000 0.00000
5 49 0.0000 0.00000
6 6 0.0000 0.00000
7 7 0.0000 0.00000
8 50 0.0000 0.00000
9 51 0.9318 0.01905
10 1 0.9990 0.10000
11 2 1.0000 0.14160
12 52 0.0000 0.00000
13 4 0.9958 0.04815
14 5 1.0000 0.08125
15 53 0.0000 0.00000
16 54 0.0000 0.00000
17 55 0.0000 0.00000
18 11 1.0000 0.25095
19 56 0.0000 0.00000
20 12 0.0000 0.00000
21 10 0.0000 0.00000
22 57 0.9895 0.02870
23 44 0.9847 0.09830
24 43 0.9980 0.09920
25 58 0.0000 0.00000
26 3 0.9784 0.02780
27 59 0.0000 0.00000
28 60 0.9780 0.10005
29 8 0.9716 0.48660
30 9 1.0000 0.42810
31 61 0.0000 0.00000
32 42 0.9423 0.02080
33 41 0.9653 0.03170
34 62 0.0000 0.00000
35 63 0.0000 0.00000
36 64 0.9923 0.03905
37 39 0.0000 0.00000
38 65 1.0000 0.01215
39 21 0.0000 0.00000
40 66 1.0000 0.02290
41 22 0.9988 0.32165
42 67 0.9846 0.03900
43 36 0.0000 0.00000
44 35 0.0000 0.00000
45 68 0.0000 0.00000
46 69 0.0000 0.00000
47 25 0.0000 0.00000
48 70 0.0000 0.00000
49 24 0.0000 0.00000
50 23 0.0000 0.00000
51 26 0.0000 0.00000
52 71 0.0000 0.00000
53 72 0.0000 0.00000
54 40 0.9855 0.03440
55 34 1.0000 0.03155
56 73 0.0000 0.00000
57 74 0.0000 0.00000
58 75 0.0000 0.00000
59 76 0.0000 0.00000
60 32 0.8537 0.01025
61 77 0.9472 0.01705
62 14 0.9994 0.15475
63 13 0.9977 0.13260
64 37 0.8816 0.02955
65 78 0.0000 0.00000
66 30 0.9990 0.28895
67 79 0.0000 0.00000
68 80 0.8462 0.01495
69 27 1.0000 0.23400
70 31 0.9980 0.25450
71 81 0.9052 0.01160
72 28 0.9974 0.11755
73 29 0.9800 0.09005
74 82 0.0000 0.00000
75 33 0.9836 0.01830
76 83 0.0000 0.00000
77 20 0.0000 0.00000
78 84 0.0000 0.00000
79 17 0.0000 0.00000
80 85 0.0000 0.00000
81 16 0.0000 0.00000
82 86 0.0000 0.00000
83 19 0.0000 0.00000
84 87 0.0000 0.00000
85 15 0.0000 0.00000
86 18 0.0000 0.00000
$desc
$desc$descendants
$desc$descendants$`60`
[1] "Callithrix_geoffroyi" "Callithrix_jacchus"
$desc$descendants$`64`
[1] "Pongo_pygmaeus" "Gorilla_gorilla" "Homo_sapiens" "Pan_troglodytes"
[5] "Pan_paniscus"
$desc$descendants$`67`
[1] "Pan_troglodytes" "Pan_paniscus"
$desc$descendants$`57`
[1] "Saimiri_sciureus" "Saimiri_oerstedii"
$desc$descendants$`66`
[1] "Homo_sapiens" "Pan_troglodytes" "Pan_paniscus"
$desc$descendants$`51`
[1] "Alouatta_palliata" "Alouatta_seniculus"
$desc$descendants$`77`
[1] "Cercocebus_torquatus" "Cercocebus_galeritus"
$desc$descendants$`80`
[1] "Macaca_arctoides" "Macaca_sinica"
$desc$descendants$`65`
[1] "Gorilla_gorilla" "Homo_sapiens" "Pan_troglodytes" "Pan_paniscus"
$desc$descendants$`81`
[1] "Macaca_fascicularis" "Macaca_mulatta"
$desc$branch.shift.probability
8 9 22 30 31 11 27 14 2
0.48660 0.42810 0.32165 0.28895 0.25450 0.25095 0.23400 0.15475 0.14160
13 28 60 1 43 44 29 5 4
0.13260 0.11755 0.10005 0.10000 0.09920 0.09830 0.09005 0.08125 0.04815
64 67 40 41 34 37 57 3 66
0.03905 0.03900 0.03440 0.03170 0.03155 0.02955 0.02870 0.02780 0.02290
42 51 33 77 80 65 81 32
0.02080 0.01905 0.01830 0.01705 0.01495 0.01215 0.01160 0.01025
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
