README.md
In hw538/cfDNAPro: cfDNAPro extracts and Visualises biological features from whole genome sequencing data of cell-free DNA
cfDNAPro 
Official tutorials
This markdown aims to provide a quick start.
For in-depth documentation, please visit: https://cfdnapro.readthedocs.io/en/latest/
Which R version to use?
As of 11 Jan 2025, some R package dependencies have not been able to support R version 4.4 yet. Please try R version 4.2 or 4.3 instead.
When installing cfDNAPro, big dependencies will be downloaded, e.g., BSgenome.Hsapiens.NCBI.GRCh38, BSgenome.Hsapiens.UCSC.hg19, BSgenome.Hsapiens.UCSC.hg38,
please set the timeout parameter by executing the following command in R console: options(timeout = 300) # Set timeout to 5 minutes (default is 60s)
Declaration
cfDNAPro is designed for research only.
Why cfDNAPro?
Unlike genomic DNA, cfDNA has specific fragmentation patterns. The ambiguous definition of "fragment length" by various alignment software is raising concerns: see page 9 footnote in SAM file format spec: https://samtools.github.io/hts-specs/SAMv1.pdf
cell-free DNA data fragmentomic analysis requires single-molecule level resolution, emphasising the importance of accurate/unbiased feature extraction. The traditional tools built for solid tissue sequencing do not consider the specific properties of cfDNA sequencing data (e.g., cfDNAs are naturally fragmented with a modal fragment size of 167bp, and di-/tri-nucleotide peaks in the length distributions). Researchers might inadvertently extract the features using a sub-optimised method.
cfDNAPro is designed to resolve this issue and standardize the cfDNA fragmentomic analysis, complying with the existing building blocks in the bioconductor R ecosystem. We wish cfDNAPro to provide a catalyst for further improvements in the implementation and development of cfDNA biomarkers and multi-modal AI for various health conditions.
Input
A paired-end sequencing bam file, with duplicates marked. (e.g., using the MarkDuplicates function from Picard).
Please do not impose any filtering on the bam files; For example, do not filter by the proper-pairs flag.
cfDNAPro filters the reads by following default criteria (You can toggle those criteria using parameters built-in readBam() function):
(1) Reads mapping qualities less than 30 were discarded;
(2) Reads must be paired. Of note, by default, cfDNAPro doesn’t impose filtration by “proper pair”;
(3) No duplicate;
(4) No secondary alignment;
(5) No supplementary alignment;
(6) No unmapped reads.
Note: remember to choose the correct genome_label, a parameter in readBam() function, based on the ref genome you used for alignment.
At the moment, it supports three different ref genomes, hg19, hg38 and hg38-NCBI, For details see readBam() R documentation by typing ?readBam in the R console or
see source code:https://github.com/hw538/cfDNAPro/blob/master/R/readBam.R
Output
cfDNAPro can extract (i.e., "quantify in a standandised and robust way") these features/bio-markers:
- fragment length
- fragment start/end/upstream/downstream motifs
- copy number variation
- single nucleotide mutation
- more...
Feature extraction depends on essential data objects/R packages in the Bioconductor ecosystem, such as Rsamtools, plyranges, GenomicAlignments, GenomeInfoDb and Biostrings.
Data engineering depends on packges in the tidyverse ecosystem, such as dplyr, and stringr.
All plots depend on ggplot2 R packge.
For issues/inquiries, please contact:
Nitzan Rosenfeld Lab admin mailbox:
Rosenfeld.LabAdmin@cruk.cam.ac.uk
Author: Haichao Wang
wanghaichao2014@gmail.com
Author: Paulius D. Mennea
paulius.mennea@cruk.cam.ac.uk
Installation
Please install our latest version(recommended):
if (!require(devtools)) install.packages("devtools")
library(devtools)
devtools::install_github("hw538/cfDNAPro", build_vignettes = TRUE, dependencies = TRUE)
# run below instead if you don't want to build vignettes inside R
# devtools::install_github("hw538/cfDNAPro", build_vignettes = FALSE, dependencies = FALSE)
Quick Start 1
Read bam file, return the fragment name (i.e. read name in bam file) and alignment coordinates in GRanges object in R.
If needed, you can convert the GRanges into a dataframe and the fragment length is stored in the "width" column.
library(cfDNAPro)
# read bam file, do alignment curation
frags <- readBam(bamfile = "/path/to/bamfile.bam")
# convert GRanges object to a dataframe in R
frag_df <- as.data.frame(frags)
# You can calculate fragment length and motifs from the frags object (i.e., the output of readBam() function)
frag_length <- callLength(frags)
frag_motif <- callMotif(frags)
A screenshot of the output:
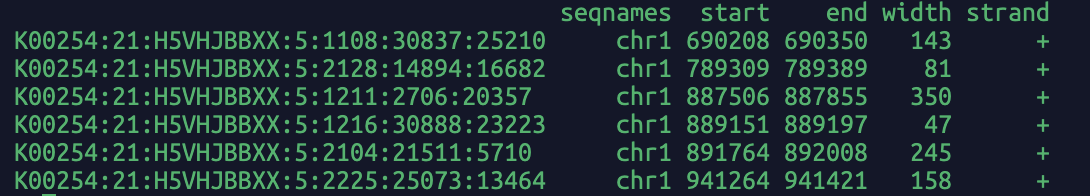
Quick Start 2
Read in bam file, return the fragment length counts.
A straightforward and frequent user case: calculate the fragment size of a bam file, use the following code:
# install cfDNAPro newest version
if (!require(devtools)) install.packages("devtools")
devtools::install_github("hw538/cfDNAPro", build_vignettes = FALSE)
# calculate insert size of a bam file
library(cfDNAPro)
frag_lengths <- read_bam_insert_metrics(bamfile = "/path/to/bamfile.bam")
The returned dataframe contains two columns, i.e., "insert_size" (fragment length) and "All_Reads.fr_count" (the count of the fragment length). A screenshot of the output:
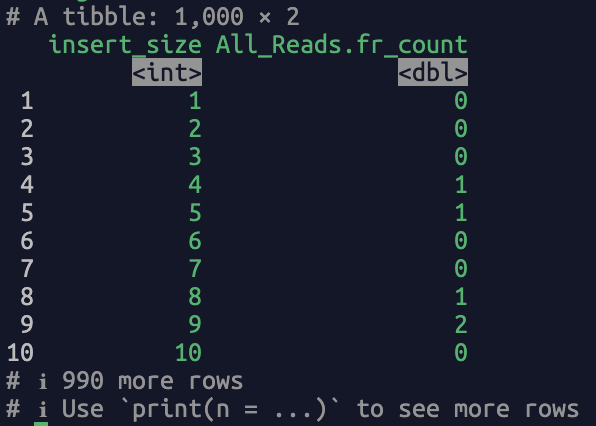
News
cfDNAPro 1.7.3 (Jan 2025)
- Updated various functions for mutational analysis
cfDNAPro 1.7.2 (Jan 2025)
- Improved various function for mutation annotation analysis etc
cfDNAPro 1.7.1 (Jan 2025)
- Improved the information and layout of this markdown quick start landing page
cfDNAPro 1.7.1 (Aug 2024)
- multiple updates
cfDNAPro 1.7.1 (May 2023)
- Resolved issues when building vignette
- Various updates
- Added/Updated readBam() functions
cfDNAPro 1.5.4 (Nov 2022)
- In addition to "bam" and "picard" files as the input, now we accept
"cfdnapro" as input_type to various functions, this 'cfdnapro' input is exactly
the output of
read_bam_insert_metrics function in cfDNAPro package. It is a
tsv file containing two columns, i.e., "insert_size" (fragment length) and
"All_Reads.fr_count" (the count of the fragment length).
cfDNAPro 1.5.3 (Oct 2022)
- added support for hg38-NCBI version, i.e. GRCh38
cfDNAPro 0.99.3 (July 2021)
- Modified vignette.
cfDNAPro 0.99.2 (July 2021)
- Modified vignette.
cfDNAPro 0.99.1 (May 2021)
- Added 'cfDNAPro' into the "watched tag".
cfDNAPro 0.99.0 (May 2021)
- Now cfDNAPro supports bam file as input for data characterisation.
- Coding style improvements.
- Documentation improvements.
- Submitted to Bioconductor.
Citation
Please cite package ‘cfDNAPro’ in publications:
Haichao Wang, Paulius Mennea, Elkie Chan, Hui Zhao, Christopher G. Smith, Tomer Kaplan, Florian Markowetz, Nitzan Rosenfeld(2024). cfDNAPro:An R/Bioconductor package to extract and visualise cell-free DNA biological features. R package version 1.7.2 https://github.com/nrlab-CRUK/cfDNAPro
hw538/cfDNAPro documentation built on April 12, 2025, 12:57 p.m.
cfDNAPro 
Official tutorials
This markdown aims to provide a quick start. For in-depth documentation, please visit: https://cfdnapro.readthedocs.io/en/latest/
Which R version to use?
As of 11 Jan 2025, some R package dependencies have not been able to support R version 4.4 yet. Please try R version 4.2 or 4.3 instead.
When installing cfDNAPro, big dependencies will be downloaded, e.g., BSgenome.Hsapiens.NCBI.GRCh38, BSgenome.Hsapiens.UCSC.hg19, BSgenome.Hsapiens.UCSC.hg38,
please set the timeout parameter by executing the following command in R console: options(timeout = 300) # Set timeout to 5 minutes (default is 60s)
Declaration
cfDNAPro is designed for research only.
Why cfDNAPro?
Unlike genomic DNA, cfDNA has specific fragmentation patterns. The ambiguous definition of "fragment length" by various alignment software is raising concerns: see page 9 footnote in SAM file format spec: https://samtools.github.io/hts-specs/SAMv1.pdf cell-free DNA data fragmentomic analysis requires single-molecule level resolution, emphasising the importance of accurate/unbiased feature extraction. The traditional tools built for solid tissue sequencing do not consider the specific properties of cfDNA sequencing data (e.g., cfDNAs are naturally fragmented with a modal fragment size of 167bp, and di-/tri-nucleotide peaks in the length distributions). Researchers might inadvertently extract the features using a sub-optimised method.
cfDNAPro is designed to resolve this issue and standardize the cfDNA fragmentomic analysis, complying with the existing building blocks in the bioconductor R ecosystem. We wish cfDNAPro to provide a catalyst for further improvements in the implementation and development of cfDNA biomarkers and multi-modal AI for various health conditions.
Input
A paired-end sequencing bam file, with duplicates marked. (e.g., using the MarkDuplicates function from Picard).
Please do not impose any filtering on the bam files; For example, do not filter by the proper-pairs flag.
cfDNAPro filters the reads by following default criteria (You can toggle those criteria using parameters built-in readBam() function):
(1) Reads mapping qualities less than 30 were discarded;
(2) Reads must be paired. Of note, by default, cfDNAPro doesn’t impose filtration by “proper pair”;
(3) No duplicate;
(4) No secondary alignment;
(5) No supplementary alignment;
(6) No unmapped reads.
Note: remember to choose the correct genome_label, a parameter in readBam() function, based on the ref genome you used for alignment.
At the moment, it supports three different ref genomes, hg19, hg38 and hg38-NCBI, For details see readBam() R documentation by typing ?readBam in the R console or
see source code:https://github.com/hw538/cfDNAPro/blob/master/R/readBam.R
Output
cfDNAPro can extract (i.e., "quantify in a standandised and robust way") these features/bio-markers:
- fragment length
- fragment start/end/upstream/downstream motifs
- copy number variation
- single nucleotide mutation
- more...
Feature extraction depends on essential data objects/R packages in the Bioconductor ecosystem, such as Rsamtools, plyranges, GenomicAlignments, GenomeInfoDb and Biostrings.
Data engineering depends on packges in the tidyverse ecosystem, such as dplyr, and stringr.
All plots depend on ggplot2 R packge.
For issues/inquiries, please contact: Nitzan Rosenfeld Lab admin mailbox: Rosenfeld.LabAdmin@cruk.cam.ac.uk Author: Haichao Wang wanghaichao2014@gmail.com Author: Paulius D. Mennea paulius.mennea@cruk.cam.ac.uk
Installation
Please install our latest version(recommended):
if (!require(devtools)) install.packages("devtools")
library(devtools)
devtools::install_github("hw538/cfDNAPro", build_vignettes = TRUE, dependencies = TRUE)
# run below instead if you don't want to build vignettes inside R
# devtools::install_github("hw538/cfDNAPro", build_vignettes = FALSE, dependencies = FALSE)
Quick Start 1
Read bam file, return the fragment name (i.e. read name in bam file) and alignment coordinates in GRanges object in R. If needed, you can convert the GRanges into a dataframe and the fragment length is stored in the "width" column.
library(cfDNAPro)
# read bam file, do alignment curation
frags <- readBam(bamfile = "/path/to/bamfile.bam")
# convert GRanges object to a dataframe in R
frag_df <- as.data.frame(frags)
# You can calculate fragment length and motifs from the frags object (i.e., the output of readBam() function)
frag_length <- callLength(frags)
frag_motif <- callMotif(frags)
A screenshot of the output:
Quick Start 2
Read in bam file, return the fragment length counts. A straightforward and frequent user case: calculate the fragment size of a bam file, use the following code:
# install cfDNAPro newest version
if (!require(devtools)) install.packages("devtools")
devtools::install_github("hw538/cfDNAPro", build_vignettes = FALSE)
# calculate insert size of a bam file
library(cfDNAPro)
frag_lengths <- read_bam_insert_metrics(bamfile = "/path/to/bamfile.bam")
The returned dataframe contains two columns, i.e., "insert_size" (fragment length) and "All_Reads.fr_count" (the count of the fragment length). A screenshot of the output:
News
cfDNAPro 1.7.3 (Jan 2025)
- Updated various functions for mutational analysis
cfDNAPro 1.7.2 (Jan 2025)
- Improved various function for mutation annotation analysis etc
cfDNAPro 1.7.1 (Jan 2025)
- Improved the information and layout of this markdown quick start landing page
cfDNAPro 1.7.1 (Aug 2024)
- multiple updates
cfDNAPro 1.7.1 (May 2023)
- Resolved issues when building vignette
- Various updates
- Added/Updated readBam() functions
cfDNAPro 1.5.4 (Nov 2022)
- In addition to "bam" and "picard" files as the input, now we accept
"cfdnapro" as input_type to various functions, this 'cfdnapro' input is exactly
the output of
read_bam_insert_metricsfunction in cfDNAPro package. It is a tsv file containing two columns, i.e., "insert_size" (fragment length) and "All_Reads.fr_count" (the count of the fragment length).
cfDNAPro 1.5.3 (Oct 2022)
- added support for hg38-NCBI version, i.e. GRCh38
cfDNAPro 0.99.3 (July 2021)
- Modified vignette.
cfDNAPro 0.99.2 (July 2021)
- Modified vignette.
cfDNAPro 0.99.1 (May 2021)
- Added 'cfDNAPro' into the "watched tag".
cfDNAPro 0.99.0 (May 2021)
- Now cfDNAPro supports bam file as input for data characterisation.
- Coding style improvements.
- Documentation improvements.
- Submitted to Bioconductor.
Citation
Please cite package ‘cfDNAPro’ in publications:
Haichao Wang, Paulius Mennea, Elkie Chan, Hui Zhao, Christopher G. Smith, Tomer Kaplan, Florian Markowetz, Nitzan Rosenfeld(2024). cfDNAPro:An R/Bioconductor package to extract and visualise cell-free DNA biological features. R package version 1.7.2 https://github.com/nrlab-CRUK/cfDNAPro
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
