README.md
In noriakis/CBNplotTest: plot bayesian network inferred from gene expression data based on enrichment analysis results
CBNplot: Bayesian network plot for the enrichment analysis results
Plot bayesian network inferred from expression data based on the enrichment analysis results from libraries including clusterProfiler and ReactomePA using bnlearn.
Installation
From Bioconductor:
BiocManager::install("CBNplot")
From GitHub:
library(devtools)
install_github("noriakis/CBNplot")
Usage
- Documentation: https://noriakis.github.io/software/CBNplot/
- Web server: https://cbnplot.com
Plot examples
-
The gene-to-gene relationship compared with the reference network.

-
The plot is customizable highliting edges and nodes like hub genes.
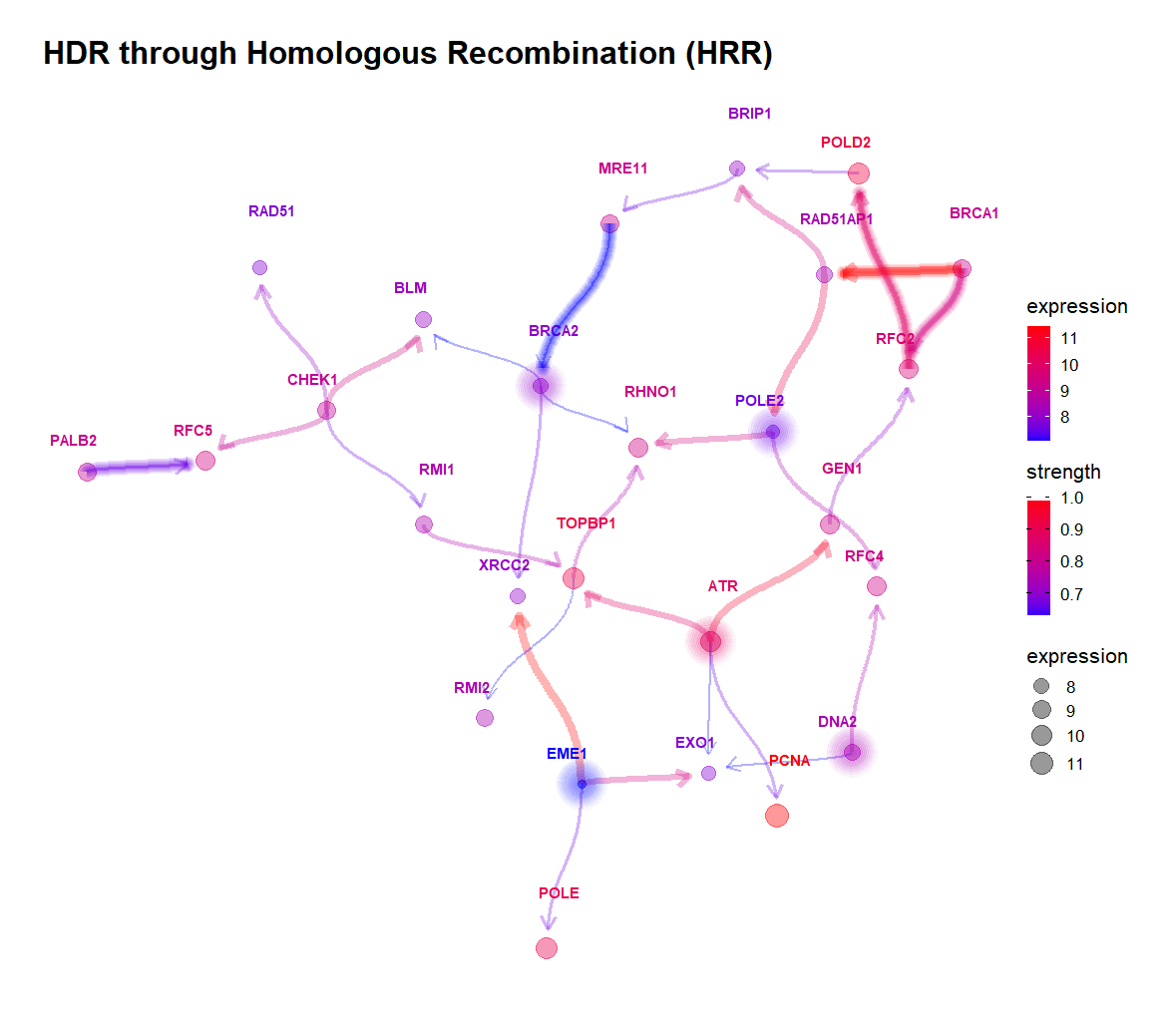
-
The example using MicrobiomeProfiler, thanks to the fix by @xiangpin.
library(MicrobiomeProfiler)]
data(Rat_data)
ko.res <- enrichKO(Rat_data)
exp.dat <- matrix(abs(rnorm(910)), 91, 10) %>% magrittr::set_rownames(value=Rat_data) %>% magrittr::set_colnames(value=paste0('S', seq_len(ncol(.))))
exp.dat %>% head
library(CBNplot)
bngeneplot(ko.res, exp=exp.dat, pathNum=1, orgDb=NULL)
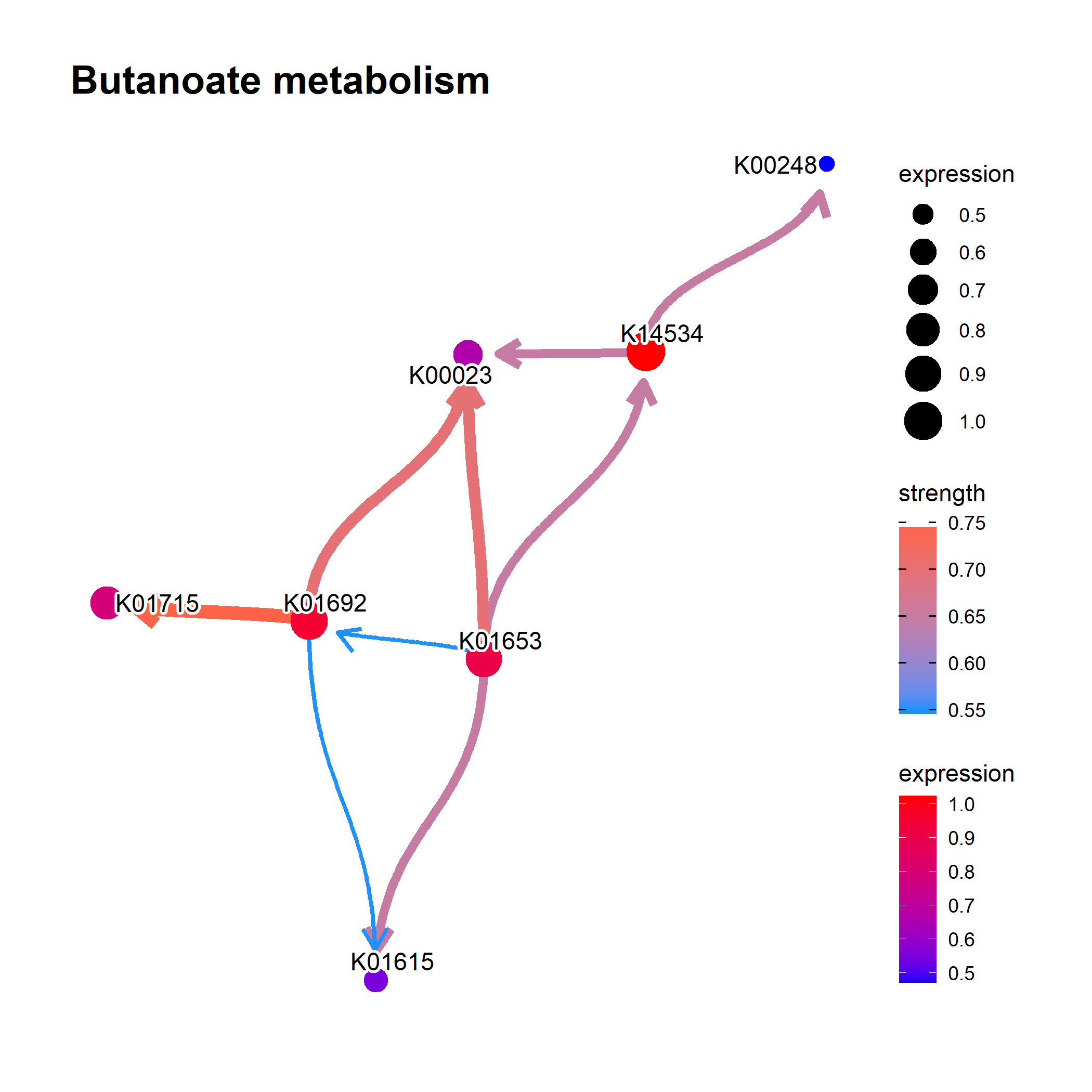
- Another customized plot.

noriakis/CBNplotTest documentation built on Nov. 18, 2022, 8:05 a.m.
CBNplot: Bayesian network plot for the enrichment analysis results
Plot bayesian network inferred from expression data based on the enrichment analysis results from libraries including clusterProfiler and ReactomePA using bnlearn.
Installation
From Bioconductor:
BiocManager::install("CBNplot")
From GitHub:
library(devtools)
install_github("noriakis/CBNplot")
Usage
- Documentation: https://noriakis.github.io/software/CBNplot/
- Web server: https://cbnplot.com
Plot examples
-
The gene-to-gene relationship compared with the reference network.

-
The plot is customizable highliting edges and nodes like hub genes.

-
The example using
MicrobiomeProfiler, thanks to the fix by @xiangpin.
library(MicrobiomeProfiler)]
data(Rat_data)
ko.res <- enrichKO(Rat_data)
exp.dat <- matrix(abs(rnorm(910)), 91, 10) %>% magrittr::set_rownames(value=Rat_data) %>% magrittr::set_colnames(value=paste0('S', seq_len(ncol(.))))
exp.dat %>% head
library(CBNplot)
bngeneplot(ko.res, exp=exp.dat, pathNum=1, orgDb=NULL)

- Another customized plot.

Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
