README.md
In data.table: Extension of `data.frame`
data.table 
data.table provides a high-performance version of base R's data.frame with syntax and feature enhancements for ease of use, convenience and programming speed.
Why data.table?
- concise syntax: fast to type, fast to read
- fast speed
- memory efficient
- careful API lifecycle management
- community
- feature rich
Features
- fast and friendly delimited file reader:
?fread, see also convenience features for small data
- fast and feature rich delimited file writer:
?fwrite
- low-level parallelism: many common operations are internally parallelized to use multiple CPU threads
- fast and scalable aggregations; e.g. 100GB in RAM (see benchmarks on up to two billion rows)
- fast and feature rich joins: ordered joins (e.g. rolling forwards, backwards, nearest and limited staleness), overlapping range joins (similar to
IRanges::findOverlaps), non-equi joins (i.e. joins using operators >, >=, <, <=), aggregate on join (by=.EACHI), update on join
- fast add/update/delete columns by reference by group using no copies at all
- fast and feature rich reshaping data:
?dcast (pivot/wider/spread) and ?melt (unpivot/longer/gather)
- any R function from any R package can be used in queries not just the subset of functions made available by a database backend, also columns of type
list are supported
- has no dependencies at all other than base R itself, for simpler production/maintenance
- the R dependency is as old as possible for as long as possible, dated April 2014, and we continuously test against that version; e.g. v1.11.0 released on 5 May 2018 bumped the dependency up from 5 year old R 3.0.0 to 4 year old R 3.1.0
Installation
install.packages("data.table")
# latest development version (only if newer available)
data.table::update_dev_pkg()
# latest development version (force install)
install.packages("data.table", repos="https://rdatatable.gitlab.io/data.table")
See the Installation wiki for more details.
Usage
Use data.table subset [ operator the same way you would use data.frame one, but...
- no need to prefix each column with
DT$ (like subset() and with() but built-in)
- any R expression using any package is allowed in
j argument, not just list of columns
- extra argument
by to compute j expression by group
library(data.table)
DT = as.data.table(iris)
# FROM[WHERE, SELECT, GROUP BY]
# DT [i, j, by]
DT[Petal.Width > 1.0, mean(Petal.Length), by = Species]
# Species V1
#1: versicolor 4.362791
#2: virginica 5.552000
Getting started
- Introduction to data.table vignette
- Getting started wiki page
- Examples produced by
example(data.table)
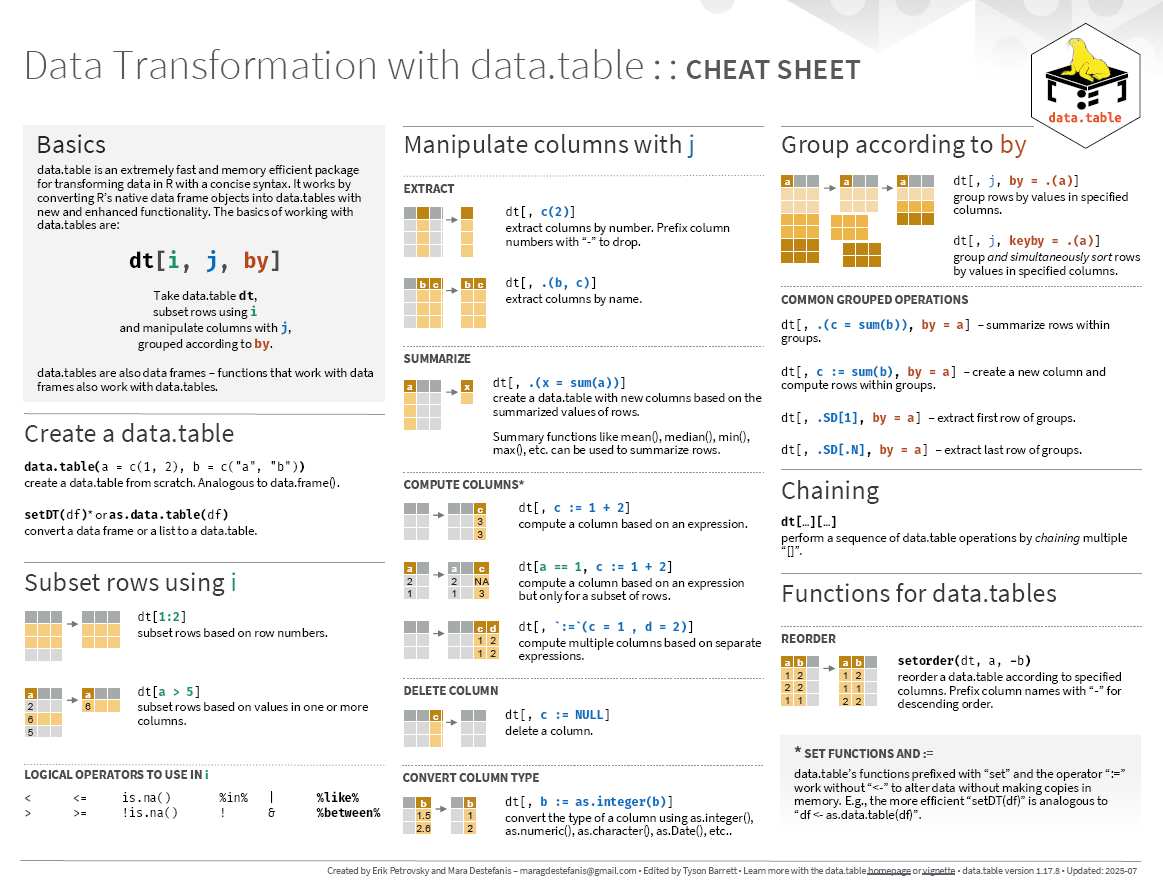
Cheatsheets
Community
data.table is widely used by the R community. It is being directly used by hundreds of CRAN and Bioconductor packages, and indirectly by thousands. It is one of the top most starred R packages on GitHub, and was highly rated by the Depsy project. If you need help, the data.table community is active on StackOverflow.
A list of packages that significantly support, extend, or make use of data.table can be found in the Seal of Approval document.
Stay up-to-date
- click the Watch button at the top and right of GitHub project page
- read NEWS file
- follow #rdatatable and the r_data_table account on X/Twitter
- follow #rdatatable and the r_data_table account on fosstodon
- follow the data.table community page on LinkedIn
- watch recent Presentations
- read recent Articles
- read posts on The Raft
Contributing
Guidelines for filing issues / pull requests: Contribution Guidelines.
Try the data.table package in your browser
Any scripts or data that you put into this service are public.
data.table documentation built on Aug. 8, 2025, 6:20 p.m.
data.table 
data.table provides a high-performance version of base R's data.frame with syntax and feature enhancements for ease of use, convenience and programming speed.
Why data.table?
- concise syntax: fast to type, fast to read
- fast speed
- memory efficient
- careful API lifecycle management
- community
- feature rich
Features
- fast and friendly delimited file reader:
?fread, see also convenience features for small data - fast and feature rich delimited file writer:
?fwrite - low-level parallelism: many common operations are internally parallelized to use multiple CPU threads
- fast and scalable aggregations; e.g. 100GB in RAM (see benchmarks on up to two billion rows)
- fast and feature rich joins: ordered joins (e.g. rolling forwards, backwards, nearest and limited staleness), overlapping range joins (similar to
IRanges::findOverlaps), non-equi joins (i.e. joins using operators>, >=, <, <=), aggregate on join (by=.EACHI), update on join - fast add/update/delete columns by reference by group using no copies at all
- fast and feature rich reshaping data:
?dcast(pivot/wider/spread) and?melt(unpivot/longer/gather) - any R function from any R package can be used in queries not just the subset of functions made available by a database backend, also columns of type
listare supported - has no dependencies at all other than base R itself, for simpler production/maintenance
- the R dependency is as old as possible for as long as possible, dated April 2014, and we continuously test against that version; e.g. v1.11.0 released on 5 May 2018 bumped the dependency up from 5 year old R 3.0.0 to 4 year old R 3.1.0
Installation
install.packages("data.table")
# latest development version (only if newer available)
data.table::update_dev_pkg()
# latest development version (force install)
install.packages("data.table", repos="https://rdatatable.gitlab.io/data.table")
See the Installation wiki for more details.
Usage
Use data.table subset [ operator the same way you would use data.frame one, but...
- no need to prefix each column with
DT$(likesubset()andwith()but built-in) - any R expression using any package is allowed in
jargument, not just list of columns - extra argument
byto computejexpression by group
library(data.table)
DT = as.data.table(iris)
# FROM[WHERE, SELECT, GROUP BY]
# DT [i, j, by]
DT[Petal.Width > 1.0, mean(Petal.Length), by = Species]
# Species V1
#1: versicolor 4.362791
#2: virginica 5.552000
Getting started
- Introduction to data.table vignette
- Getting started wiki page
- Examples produced by
example(data.table)
Cheatsheets
Community
data.table is widely used by the R community. It is being directly used by hundreds of CRAN and Bioconductor packages, and indirectly by thousands. It is one of the top most starred R packages on GitHub, and was highly rated by the Depsy project. If you need help, the data.table community is active on StackOverflow.
A list of packages that significantly support, extend, or make use of data.table can be found in the Seal of Approval document.
Stay up-to-date
- click the Watch button at the top and right of GitHub project page
- read NEWS file
- follow #rdatatable and the r_data_table account on X/Twitter
- follow #rdatatable and the r_data_table account on fosstodon
- follow the data.table community page on LinkedIn
- watch recent Presentations
- read recent Articles
- read posts on The Raft
Contributing
Guidelines for filing issues / pull requests: Contribution Guidelines.
Try the data.table package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.