README.md
In phac-nml-phrsd/angedist: Antigenic Genetic Distance
angedist
Antigenic Genetic Distance
Warning: this repo is a work in progress
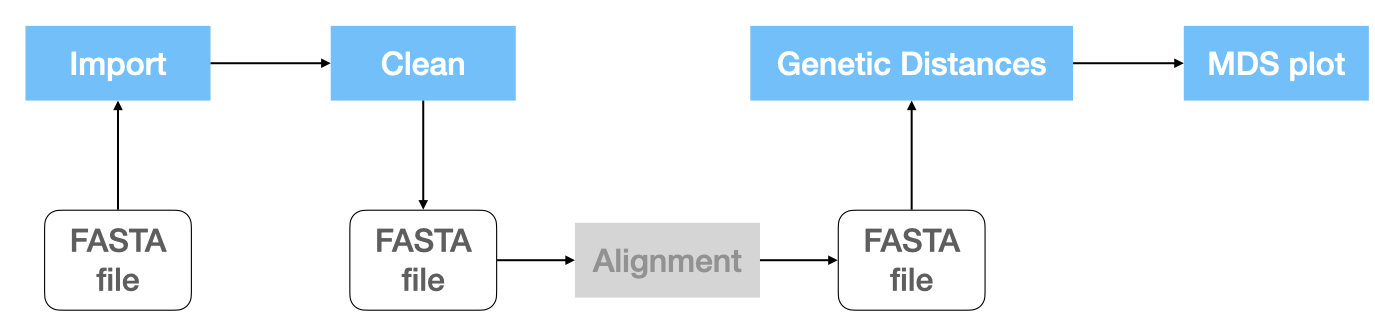
Pipeline
Installation
devtools::install_github('phac-nml-phrsd/angedist')
Simple examples
There are a few FASTA files attached to the angedist package.
Let's use them in simple examples
To import a FASTA file, use the function import_seqs().
library(angedist)
path1 = system.file("extdata", "example1.fasta", package = "angedist")
seqs = import_seqs(path1,
prms = list(seq.source = 'GISAID',
pathogen = 'H3N2',
seq.type = 'AA'))
Given that we are working with influenza sequences, we can filter out low-quality sequences using clean_seq_influenza_A()
seqs.cleaned = clean_seq_influenza_A(seqs, verbose = TRUE)
TO DO DOCUMENT THE CODE BELOW
seqs.aligned = import_seqs('H3N2_cleaned_align.fasta',
prms = list(seq.source = 'GISAID',
pathogen = 'H3N2',
seq.type = 'AA'))
m = dist_matrix(sobj = seqs.aligned,
sites = NULL,
ncores = 4,
dist.type = 'hamming')
metavars = list(sname = seqs.aligned$strain.name,
datec = seqs.aligned$date.collection)
mdsobj = mds(m, dim.mds = 2, metavars=metavars)
mdsobj2 = mds(m, dim.mds = 2, metavars=NULL)
# highlight sequence from Aichi
mdsobj$df$highlight = grepl('Aichi', mdsobj$df$sname)
mdsobj$df$highlight.label = ifelse(mdsobj$df$highlight,'Aichi','')
g = plot_mds(mdsobj = mdsobj, color_varname = 'datec',
highlight = TRUE, highlight.label = TRUE)
g2 = plot_mds(mdsobj = mdsobj)
plot(g)
plot(g2)
phac-nml-phrsd/angedist documentation built on Nov. 27, 2022, 7:23 p.m.
angedist
Antigenic Genetic Distance
Warning: this repo is a work in progress
Pipeline

Installation
devtools::install_github('phac-nml-phrsd/angedist')
Simple examples
There are a few FASTA files attached to the angedist package.
Let's use them in simple examples
To import a FASTA file, use the function import_seqs().
library(angedist)
path1 = system.file("extdata", "example1.fasta", package = "angedist")
seqs = import_seqs(path1,
prms = list(seq.source = 'GISAID',
pathogen = 'H3N2',
seq.type = 'AA'))
Given that we are working with influenza sequences, we can filter out low-quality sequences using clean_seq_influenza_A()
seqs.cleaned = clean_seq_influenza_A(seqs, verbose = TRUE)
TO DO DOCUMENT THE CODE BELOW
seqs.aligned = import_seqs('H3N2_cleaned_align.fasta',
prms = list(seq.source = 'GISAID',
pathogen = 'H3N2',
seq.type = 'AA'))
m = dist_matrix(sobj = seqs.aligned,
sites = NULL,
ncores = 4,
dist.type = 'hamming')
metavars = list(sname = seqs.aligned$strain.name,
datec = seqs.aligned$date.collection)
mdsobj = mds(m, dim.mds = 2, metavars=metavars)
mdsobj2 = mds(m, dim.mds = 2, metavars=NULL)
# highlight sequence from Aichi
mdsobj$df$highlight = grepl('Aichi', mdsobj$df$sname)
mdsobj$df$highlight.label = ifelse(mdsobj$df$highlight,'Aichi','')
g = plot_mds(mdsobj = mdsobj, color_varname = 'datec',
highlight = TRUE, highlight.label = TRUE)
g2 = plot_mds(mdsobj = mdsobj)
plot(g)
plot(g2)
Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
