serially: Run several tests and a final validation in a serial manner
In pointblank: Data Validation and Organization of Metadata for Local and Remote Tables
serially R Documentation
Run several tests and a final validation in a serial manner
Description
The serially() validation function allows for a series of tests to run in
sequence before either culminating in a final validation step or simply
exiting the series. This construction allows for pre-testing that may make
sense before a validation step. For example, there may be situations where
it's vital to check a column type before performing a validation on the same
column (since having the wrong type can result in an evaluation error for the
subsequent validation). Another serial workflow might entail having a bundle
of checks in a prescribed order and, if all pass, then the goal of this
testing has been achieved (e.g., checking if a table matches another through
a series of increasingly specific tests).
A series as specified inside serially() is composed with a listing of
calls, and we would draw upon test functions (T) to describe tests and
optionally provide a finalizing call with a validation function (V).
The following constraints apply:
there must be at least one test function in the series (T -> V is
good, V is not)
there can only be one validation function call, V; it's optional but,
if included, it must be placed at the end (T -> T -> V is good,
these sequences are bad: (1) T -> V -> T, (2) T -> T ->
V -> V)
a validation function call (V), if included, mustn't itself yield
multiple validation steps (this may happen when providing multiple columns
or any segments)
Here's an example of how to arrange expressions:
~ test_col_exists(., columns = count),
~ test_col_is_numeric(., columns = count),
~ col_vals_gt(., columns = count, value = 2)
This series concentrates on the column called count and first checks
whether the column exists, then checks if that column is numeric, and then
finally validates whether all values in the column are greater than 2.
Note that in the above listing of calls, the . stands in for the target
table and is always necessary here. Also important is that all test_*()
functions have a threshold argument that is set to 1 by default. Should
you need to bump up the threshold value it can be changed to a different
integer value (as an absolute threshold of failing test units) or a
decimal value between 0 and 1 (serving as a fractional threshold of
failing test units).
Usage
serially(
x,
...,
.list = list2(...),
preconditions = NULL,
actions = NULL,
step_id = NULL,
label = NULL,
brief = NULL,
active = TRUE
)
expect_serially(
object,
...,
.list = list2(...),
preconditions = NULL,
threshold = 1
)
test_serially(
object,
...,
.list = list2(...),
preconditions = NULL,
threshold = 1
)
Arguments
x
A pointblank agent or a data table
obj:<ptblank_agent>|obj:<tbl_*> // required
A data frame, tibble (tbl_df or tbl_dbi), Spark DataFrame
(tbl_spark), or, an agent object of class ptblank_agent that is
commonly created with create_agent().
...
Test/validation expressions
<test/validation expressions> // required (or, use .list)
A collection one-sided formulas that consist of test_*() function calls
(e.g., test_col_vals_between(), etc.) arranged in sequence of intended
interrogation order. Typically, validations up until the final one would
have some threshold value set (default is 1) for short circuiting
within the series. A finishing validation function call (e.g.,
col_vals_increasing(), etc.) can optionally be inserted at the end of the
series, serving as a validation step that only undergoes interrogation if
the prior tests adequately pass. An example of this is
~ test_column_exists(., a), ~ col_vals_not_null(., a)).
.list
Alternative to ...
<list of multiple expressions> // required (or, use ...)
Allows for the use of a list as an input alternative to ....
preconditions
Input table modification prior to validation
<table mutation expression> // default: NULL (optional)
An optional expression for mutating the input table before proceeding with
the validation. This can either be provided as a one-sided R formula using
a leading ~ (e.g., ~ . %>% dplyr::mutate(col = col + 10) or as a
function (e.g., function(x) dplyr::mutate(x, col = col + 10). See the
Preconditions section for more information.
actions
Thresholds and actions for different states
obj:<action_levels> // default: NULL (optional)
A list containing threshold levels so that the validation step can react
accordingly when exceeding the set levels for different states. This is to
be created with the action_levels() helper function.
step_id
Manual setting of the step ID value
scalar<character> // default: NULL (optional)
One or more optional identifiers for the single or multiple validation
steps generated from calling a validation function. The use of step IDs
serves to distinguish validation steps from each other and provide an
opportunity for supplying a more meaningful label compared to the step
index. By default this is NULL, and pointblank will automatically
generate the step ID value (based on the step index) in this case. One or
more values can be provided, and the exact number of ID values should (1)
match the number of validation steps that the validation function call will
produce (influenced by the number of columns provided), (2) be an ID
string not used in any previous validation step, and (3) be a vector with
unique values.
label
Optional label for the validation step
vector<character> // default: NULL (optional)
Optional label for the validation step. This label appears in the agent
report and, for the best appearance, it should be kept quite short. See
the Labels section for more information.
brief
Brief description for the validation step
scalar<character> // default: NULL (optional)
A brief is a short, text-based description for the validation step. If
nothing is provided here then an autobrief is generated by the agent,
using the language provided in create_agent()'s lang argument (which
defaults to "en" or English). The autobrief incorporates details of the
validation step so it's often the preferred option in most cases (where a
label might be better suited to succinctly describe the validation).
active
Is the validation step active?
scalar<logical> // default: TRUE
A logical value indicating whether the validation step should be active. If
the validation function is working with an agent, FALSE will make the
validation step inactive (still reporting its presence and keeping indexes
for the steps unchanged). If the validation function will be operating
directly on data (no agent involvement), then any step with active = FALSE will simply pass the data through with no validation whatsoever.
Aside from a logical vector, a one-sided R formula using a leading ~ can
be used with . (serving as the input data table) to evaluate to a single
logical value. With this approach, the pointblank function
has_columns() can be used to determine whether to make a validation step
active on the basis of one or more columns existing in the table
(e.g., ~ . %>% has_columns(c(d, e))).
object
A data table for expectations or tests
obj:<tbl_*> // required
A data frame, tibble (tbl_df or tbl_dbi), or Spark DataFrame
(tbl_spark) that serves as the target table for the expectation function
or the test function.
threshold
The failure threshold
scalar<integer|numeric>(val>=0) // default: 1
A simple failure threshold value for use with the expectation (expect_)
and the test (test_) function variants. By default, this is set to 1
meaning that any single unit of failure in data validation results in an
overall test failure. Whole numbers beyond 1 indicate that any failing
units up to that absolute threshold value will result in a succeeding
testthat test or evaluate to TRUE. Likewise, fractional values
(between 0 and 1) act as a proportional failure threshold, where 0.15
means that 15 percent of failing test units results in an overall test
failure.
Value
For the validation function, the return value is either a
ptblank_agent object or a table object (depending on whether an agent
object or a table was passed to x). The expectation function invisibly
returns its input but, in the context of testing data, the function is
called primarily for its potential side-effects (e.g., signaling failure).
The test function returns a logical value.
Supported Input Tables
The types of data tables that are officially supported are:
data frames (data.frame) and tibbles (tbl_df)
Spark DataFrames (tbl_spark)
the following database tables (tbl_dbi):
-
PostgreSQL tables (using the RPostgres::Postgres() as driver)
-
MySQL tables (with RMySQL::MySQL())
-
Microsoft SQL Server tables (via odbc)
-
BigQuery tables (using bigrquery::bigquery())
-
DuckDB tables (through duckdb::duckdb())
-
SQLite (with RSQLite::SQLite())
Other database tables may work to varying degrees but they haven't been
formally tested (so be mindful of this when using unsupported backends with
pointblank).
Column Names
columns may be a single column (as symbol a or string "a") or a vector
of columns (c(a, b, c) or c("a", "b", "c")). {tidyselect} helpers
are also supported, such as contains("date") and where(is.double). If
passing an external vector of columns, it should be wrapped in all_of().
When multiple columns are selected by columns, the result will be an
expansion of validation steps to that number of columns (e.g.,
c(col_a, col_b) will result in the entry of two validation steps).
Previously, columns could be specified in vars(). This continues to work,
but c() offers the same capability and supersedes vars() in columns.
Preconditions
Providing expressions as preconditions means pointblank will preprocess
the target table during interrogation as a preparatory step. It might happen
that a particular validation requires a calculated column, some filtering of
rows, or the addition of columns via a join, etc. Especially for an
agent-based report this can be advantageous since we can develop a large
validation plan with a single target table and make minor adjustments to it,
as needed, along the way.
The table mutation is totally isolated in scope to the validation step(s)
where preconditions is used. Using dplyr code is suggested here since
the statements can be translated to SQL if necessary (i.e., if the target
table resides in a database). The code is most easily supplied as a one-sided
R formula (using a leading ~). In the formula representation, the .
serves as the input data table to be transformed (e.g., ~ . %>% dplyr::mutate(col_b = col_a + 10)). Alternatively, a function could instead
be supplied (e.g., function(x) dplyr::mutate(x, col_b = col_a + 10)).
Actions
Often, we will want to specify actions for the validation. This argument,
present in every validation function, takes a specially-crafted list
object that is best produced by the action_levels() function. Read that
function's documentation for the lowdown on how to create reactions to
above-threshold failure levels in validation. The basic gist is that you'll
want at least a single threshold level (specified as either the fraction of
test units failed, or, an absolute value), often using the warn_at
argument. This is especially true when x is a table object because,
otherwise, nothing happens. For the col_vals_*()-type functions, using
action_levels(warn_at = 0.25) or action_levels(stop_at = 0.25) are good
choices depending on the situation (the first produces a warning when a
quarter of the total test units fails, the other stop()s at the same
threshold level).
Labels
label may be a single string or a character vector that matches the number
of expanded steps. label also supports {glue} syntax and exposes the
following dynamic variables contextualized to the current step:
-
"{.step}": The validation step name
The glue context also supports ordinary expressions for further flexibility
(e.g., "{toupper(.step)}") as long as they return a length-1 string.
Briefs
Want to describe this validation step in some detail? Keep in mind that this
is only useful if x is an agent. If that's the case, brief the agent
with some text that fits. Don't worry if you don't want to do it. The
autobrief protocol is kicked in when brief = NULL and a simple brief will
then be automatically generated.
YAML
A pointblank agent can be written to YAML with yaml_write() and the
resulting YAML can be used to regenerate an agent (with yaml_read_agent())
or interrogate the target table (via yaml_agent_interrogate()). When
serially() is represented in YAML (under the top-level steps key as a
list member), the syntax closely follows the signature of the validation
function. Here is an example of how a complex call of serially() as a
validation step is expressed in R code and in the corresponding YAML
representation.
R statement:
agent %>%
serially(
~ test_col_vals_lt(., columns = a, value = 8),
~ test_col_vals_gt(., columns = c, value = vars(a)),
~ col_vals_not_null(., columns = b),
preconditions = ~ . %>% dplyr::filter(a < 10),
actions = action_levels(warn_at = 0.1, stop_at = 0.2),
label = "The `serially()` step.",
active = FALSE
)
YAML representation:
steps:
- serially:
fns:
- ~test_col_vals_lt(., columns = a, value = 8)
- ~test_col_vals_gt(., columns = c, value = vars(a))
- ~col_vals_not_null(., columns = b)
preconditions: ~. %>% dplyr::filter(a < 10)
actions:
warn_fraction: 0.1
stop_fraction: 0.2
label: The `serially()` step.
active: false
In practice, both of these will often be shorter as only the expressions for
validation steps are necessary. Arguments with default values won't be
written to YAML when using yaml_write() (though it is acceptable to include
them with their default when generating the YAML by other means). It is also
possible to preview the transformation of an agent to YAML without any
writing to disk by using the yaml_agent_string() function.
Examples
For all examples here, we'll use a simple table with three numeric columns
(a, b, and c). This is a very basic table but it'll be more useful when
explaining things later.
tbl <-
dplyr::tibble(
a = c(5, 2, 6),
b = c(6, 4, 9),
c = c(1, 2, 3)
)
tbl
#> # A tibble: 3 x 3
#> a b c
#> <dbl> <dbl> <dbl>
#> 1 5 6 1
#> 2 2 4 2
#> 3 6 9 3
A: Using an agent with validation functions and then interrogate()
The serially() function can be set up to perform a series of tests and then
perform a validation (only if all tests pass). Here, we are going to (1) test
whether columns a and b are numeric, (2) check that both don't have any
NA values, and (3) perform a finalizing validation that checks whether
values in b are greater than values in a. We'll determine if this
validation has any failing test units (there are 4 tests and a final
validation).
agent_1 <-
create_agent(tbl = tbl) %>%
serially(
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b)),
~ col_vals_gt(., columns = b, value = vars(a))
) %>%
interrogate()
Printing the agent in the console shows the validation report in the
Viewer. Here is an excerpt of validation report, showing the single entry
that corresponds to the validation step demonstrated here.

What's going on? All four of the tests passed and so the final validation
occurred. There were no failing test units in that either!
The final validation is optional and so here is a variation where only the
serial tests are performed.
agent_2 <-
create_agent(tbl = tbl) %>%
serially(
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b))
) %>%
interrogate()
Everything is good here too:
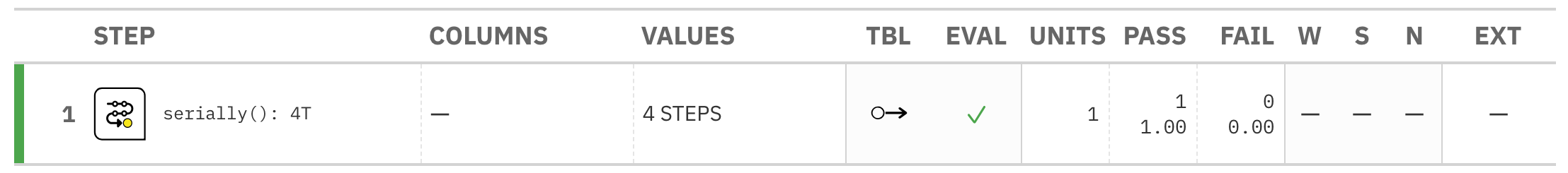
B: Using the validation function directly on the data (no agent)
This way of using validation functions acts as a data filter. Data is passed
through but should stop() if there is a single test unit failing. The
behavior of side effects can be customized with the actions option.
tbl %>%
serially(
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b)),
~ col_vals_gt(., columns = b, value = vars(a))
)
#> # A tibble: 3 x 3
#> a b c
#> <dbl> <dbl> <dbl>
#> 1 5 6 1
#> 2 2 4 2
#> 3 6 9 3
C: Using the expectation function
With the expect_*() form, we would typically perform one validation at a
time. This is primarily used in testthat tests.
expect_serially(
tbl,
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b)),
~ col_vals_gt(., columns = b, value = vars(a))
)
D: Using the test function
With the test_*() form, we should get a single logical value returned to
us.
tbl %>%
test_serially(
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b)),
~ col_vals_gt(., columns = b, value = vars(a))
)
#> [1] TRUE
Function ID
2-35
See Also
Other validation functions:
col_count_match(),
col_exists(),
col_is_character(),
col_is_date(),
col_is_factor(),
col_is_integer(),
col_is_logical(),
col_is_numeric(),
col_is_posix(),
col_schema_match(),
col_vals_between(),
col_vals_decreasing(),
col_vals_equal(),
col_vals_expr(),
col_vals_gt(),
col_vals_gte(),
col_vals_in_set(),
col_vals_increasing(),
col_vals_lt(),
col_vals_lte(),
col_vals_make_set(),
col_vals_make_subset(),
col_vals_not_between(),
col_vals_not_equal(),
col_vals_not_in_set(),
col_vals_not_null(),
col_vals_null(),
col_vals_regex(),
col_vals_within_spec(),
conjointly(),
row_count_match(),
rows_complete(),
rows_distinct(),
specially(),
tbl_match()
pointblank documentation built on Nov. 29, 2025, 1:06 a.m.
| serially | R Documentation |
Run several tests and a final validation in a serial manner
Description
The serially() validation function allows for a series of tests to run in
sequence before either culminating in a final validation step or simply
exiting the series. This construction allows for pre-testing that may make
sense before a validation step. For example, there may be situations where
it's vital to check a column type before performing a validation on the same
column (since having the wrong type can result in an evaluation error for the
subsequent validation). Another serial workflow might entail having a bundle
of checks in a prescribed order and, if all pass, then the goal of this
testing has been achieved (e.g., checking if a table matches another through
a series of increasingly specific tests).
A series as specified inside serially() is composed with a listing of
calls, and we would draw upon test functions (T) to describe tests and
optionally provide a finalizing call with a validation function (V).
The following constraints apply:
there must be at least one test function in the series (T -> V is good, V is not)
there can only be one validation function call, V; it's optional but, if included, it must be placed at the end (T -> T -> V is good, these sequences are bad: (1) T -> V -> T, (2) T -> T -> V -> V)
a validation function call (V), if included, mustn't itself yield multiple validation steps (this may happen when providing multiple
columnsor anysegments)
Here's an example of how to arrange expressions:
~ test_col_exists(., columns = count), ~ test_col_is_numeric(., columns = count), ~ col_vals_gt(., columns = count, value = 2)
This series concentrates on the column called count and first checks
whether the column exists, then checks if that column is numeric, and then
finally validates whether all values in the column are greater than 2.
Note that in the above listing of calls, the . stands in for the target
table and is always necessary here. Also important is that all test_*()
functions have a threshold argument that is set to 1 by default. Should
you need to bump up the threshold value it can be changed to a different
integer value (as an absolute threshold of failing test units) or a
decimal value between 0 and 1 (serving as a fractional threshold of
failing test units).
Usage
serially(
x,
...,
.list = list2(...),
preconditions = NULL,
actions = NULL,
step_id = NULL,
label = NULL,
brief = NULL,
active = TRUE
)
expect_serially(
object,
...,
.list = list2(...),
preconditions = NULL,
threshold = 1
)
test_serially(
object,
...,
.list = list2(...),
preconditions = NULL,
threshold = 1
)
Arguments
x |
A pointblank agent or a data table
A data frame, tibble ( |
... |
Test/validation expressions
A collection one-sided formulas that consist of |
.list |
Alternative to
Allows for the use of a list as an input alternative to |
preconditions |
Input table modification prior to validation
An optional expression for mutating the input table before proceeding with
the validation. This can either be provided as a one-sided R formula using
a leading |
actions |
Thresholds and actions for different states
A list containing threshold levels so that the validation step can react
accordingly when exceeding the set levels for different states. This is to
be created with the |
step_id |
Manual setting of the step ID value
One or more optional identifiers for the single or multiple validation
steps generated from calling a validation function. The use of step IDs
serves to distinguish validation steps from each other and provide an
opportunity for supplying a more meaningful label compared to the step
index. By default this is |
label |
Optional label for the validation step
Optional label for the validation step. This label appears in the agent report and, for the best appearance, it should be kept quite short. See the Labels section for more information. |
brief |
Brief description for the validation step
A brief is a short, text-based description for the validation step. If
nothing is provided here then an autobrief is generated by the agent,
using the language provided in |
active |
Is the validation step active?
A logical value indicating whether the validation step should be active. If
the validation function is working with an agent, |
object |
A data table for expectations or tests
A data frame, tibble ( |
threshold |
The failure threshold
A simple failure threshold value for use with the expectation ( |
Value
For the validation function, the return value is either a
ptblank_agent object or a table object (depending on whether an agent
object or a table was passed to x). The expectation function invisibly
returns its input but, in the context of testing data, the function is
called primarily for its potential side-effects (e.g., signaling failure).
The test function returns a logical value.
Supported Input Tables
The types of data tables that are officially supported are:
data frames (
data.frame) and tibbles (tbl_df)Spark DataFrames (
tbl_spark)the following database tables (
tbl_dbi):-
PostgreSQL tables (using the
RPostgres::Postgres()as driver) -
MySQL tables (with
RMySQL::MySQL()) -
Microsoft SQL Server tables (via odbc)
-
BigQuery tables (using
bigrquery::bigquery()) -
DuckDB tables (through
duckdb::duckdb()) -
SQLite (with
RSQLite::SQLite())
-
Other database tables may work to varying degrees but they haven't been formally tested (so be mindful of this when using unsupported backends with pointblank).
Column Names
columns may be a single column (as symbol a or string "a") or a vector
of columns (c(a, b, c) or c("a", "b", "c")). {tidyselect} helpers
are also supported, such as contains("date") and where(is.double). If
passing an external vector of columns, it should be wrapped in all_of().
When multiple columns are selected by columns, the result will be an
expansion of validation steps to that number of columns (e.g.,
c(col_a, col_b) will result in the entry of two validation steps).
Previously, columns could be specified in vars(). This continues to work,
but c() offers the same capability and supersedes vars() in columns.
Preconditions
Providing expressions as preconditions means pointblank will preprocess
the target table during interrogation as a preparatory step. It might happen
that a particular validation requires a calculated column, some filtering of
rows, or the addition of columns via a join, etc. Especially for an
agent-based report this can be advantageous since we can develop a large
validation plan with a single target table and make minor adjustments to it,
as needed, along the way.
The table mutation is totally isolated in scope to the validation step(s)
where preconditions is used. Using dplyr code is suggested here since
the statements can be translated to SQL if necessary (i.e., if the target
table resides in a database). The code is most easily supplied as a one-sided
R formula (using a leading ~). In the formula representation, the .
serves as the input data table to be transformed (e.g., ~ . %>% dplyr::mutate(col_b = col_a + 10)). Alternatively, a function could instead
be supplied (e.g., function(x) dplyr::mutate(x, col_b = col_a + 10)).
Actions
Often, we will want to specify actions for the validation. This argument,
present in every validation function, takes a specially-crafted list
object that is best produced by the action_levels() function. Read that
function's documentation for the lowdown on how to create reactions to
above-threshold failure levels in validation. The basic gist is that you'll
want at least a single threshold level (specified as either the fraction of
test units failed, or, an absolute value), often using the warn_at
argument. This is especially true when x is a table object because,
otherwise, nothing happens. For the col_vals_*()-type functions, using
action_levels(warn_at = 0.25) or action_levels(stop_at = 0.25) are good
choices depending on the situation (the first produces a warning when a
quarter of the total test units fails, the other stop()s at the same
threshold level).
Labels
label may be a single string or a character vector that matches the number
of expanded steps. label also supports {glue} syntax and exposes the
following dynamic variables contextualized to the current step:
-
"{.step}": The validation step name
The glue context also supports ordinary expressions for further flexibility
(e.g., "{toupper(.step)}") as long as they return a length-1 string.
Briefs
Want to describe this validation step in some detail? Keep in mind that this
is only useful if x is an agent. If that's the case, brief the agent
with some text that fits. Don't worry if you don't want to do it. The
autobrief protocol is kicked in when brief = NULL and a simple brief will
then be automatically generated.
YAML
A pointblank agent can be written to YAML with yaml_write() and the
resulting YAML can be used to regenerate an agent (with yaml_read_agent())
or interrogate the target table (via yaml_agent_interrogate()). When
serially() is represented in YAML (under the top-level steps key as a
list member), the syntax closely follows the signature of the validation
function. Here is an example of how a complex call of serially() as a
validation step is expressed in R code and in the corresponding YAML
representation.
R statement:
agent %>%
serially(
~ test_col_vals_lt(., columns = a, value = 8),
~ test_col_vals_gt(., columns = c, value = vars(a)),
~ col_vals_not_null(., columns = b),
preconditions = ~ . %>% dplyr::filter(a < 10),
actions = action_levels(warn_at = 0.1, stop_at = 0.2),
label = "The `serially()` step.",
active = FALSE
)
YAML representation:
steps:
- serially:
fns:
- ~test_col_vals_lt(., columns = a, value = 8)
- ~test_col_vals_gt(., columns = c, value = vars(a))
- ~col_vals_not_null(., columns = b)
preconditions: ~. %>% dplyr::filter(a < 10)
actions:
warn_fraction: 0.1
stop_fraction: 0.2
label: The `serially()` step.
active: false
In practice, both of these will often be shorter as only the expressions for
validation steps are necessary. Arguments with default values won't be
written to YAML when using yaml_write() (though it is acceptable to include
them with their default when generating the YAML by other means). It is also
possible to preview the transformation of an agent to YAML without any
writing to disk by using the yaml_agent_string() function.
Examples
For all examples here, we'll use a simple table with three numeric columns
(a, b, and c). This is a very basic table but it'll be more useful when
explaining things later.
tbl <-
dplyr::tibble(
a = c(5, 2, 6),
b = c(6, 4, 9),
c = c(1, 2, 3)
)
tbl
#> # A tibble: 3 x 3
#> a b c
#> <dbl> <dbl> <dbl>
#> 1 5 6 1
#> 2 2 4 2
#> 3 6 9 3
A: Using an agent with validation functions and then interrogate()
The serially() function can be set up to perform a series of tests and then
perform a validation (only if all tests pass). Here, we are going to (1) test
whether columns a and b are numeric, (2) check that both don't have any
NA values, and (3) perform a finalizing validation that checks whether
values in b are greater than values in a. We'll determine if this
validation has any failing test units (there are 4 tests and a final
validation).
agent_1 <-
create_agent(tbl = tbl) %>%
serially(
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b)),
~ col_vals_gt(., columns = b, value = vars(a))
) %>%
interrogate()
Printing the agent in the console shows the validation report in the
Viewer. Here is an excerpt of validation report, showing the single entry
that corresponds to the validation step demonstrated here.

What's going on? All four of the tests passed and so the final validation occurred. There were no failing test units in that either!
The final validation is optional and so here is a variation where only the serial tests are performed.
agent_2 <-
create_agent(tbl = tbl) %>%
serially(
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b))
) %>%
interrogate()
Everything is good here too:

B: Using the validation function directly on the data (no agent)
This way of using validation functions acts as a data filter. Data is passed
through but should stop() if there is a single test unit failing. The
behavior of side effects can be customized with the actions option.
tbl %>%
serially(
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b)),
~ col_vals_gt(., columns = b, value = vars(a))
)
#> # A tibble: 3 x 3
#> a b c
#> <dbl> <dbl> <dbl>
#> 1 5 6 1
#> 2 2 4 2
#> 3 6 9 3
C: Using the expectation function
With the expect_*() form, we would typically perform one validation at a
time. This is primarily used in testthat tests.
expect_serially( tbl, ~ test_col_is_numeric(., columns = c(a, b)), ~ test_col_vals_not_null(., columns = c(a, b)), ~ col_vals_gt(., columns = b, value = vars(a)) )
D: Using the test function
With the test_*() form, we should get a single logical value returned to
us.
tbl %>%
test_serially(
~ test_col_is_numeric(., columns = c(a, b)),
~ test_col_vals_not_null(., columns = c(a, b)),
~ col_vals_gt(., columns = b, value = vars(a))
)
#> [1] TRUE
Function ID
2-35
See Also
Other validation functions:
col_count_match(),
col_exists(),
col_is_character(),
col_is_date(),
col_is_factor(),
col_is_integer(),
col_is_logical(),
col_is_numeric(),
col_is_posix(),
col_schema_match(),
col_vals_between(),
col_vals_decreasing(),
col_vals_equal(),
col_vals_expr(),
col_vals_gt(),
col_vals_gte(),
col_vals_in_set(),
col_vals_increasing(),
col_vals_lt(),
col_vals_lte(),
col_vals_make_set(),
col_vals_make_subset(),
col_vals_not_between(),
col_vals_not_equal(),
col_vals_not_in_set(),
col_vals_not_null(),
col_vals_null(),
col_vals_regex(),
col_vals_within_spec(),
conjointly(),
row_count_match(),
rows_complete(),
rows_distinct(),
specially(),
tbl_match()
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
