README.md
In afitz-gmu/DGE-Analysis: Bioinformatics plots for various Visualization
DGE-Analysis
Index
Install
Install the required packages and link to the correct github repository.
library(remotes)
remotes::install_github(repo="afitz-gmu/DGE-Analysis", build_opts = c("--no-resave-data", "--no-manual"))
library(GenomicVisualization)
DGE
Basic function to perfrom Differential Gene Expression by EdgeR, DESeq2 and Voom.
data("count.table")
design <- data.frame("trt" = colnames(count.table))
rownames(design) <- design$trt
design$trt <- as.integer(grepl("T",design[,1]))
DGE.list <- DGE(count.table = count.table , design.matrix = design , method=c("EdgeR","voom") )
VennDig(DGE.list)
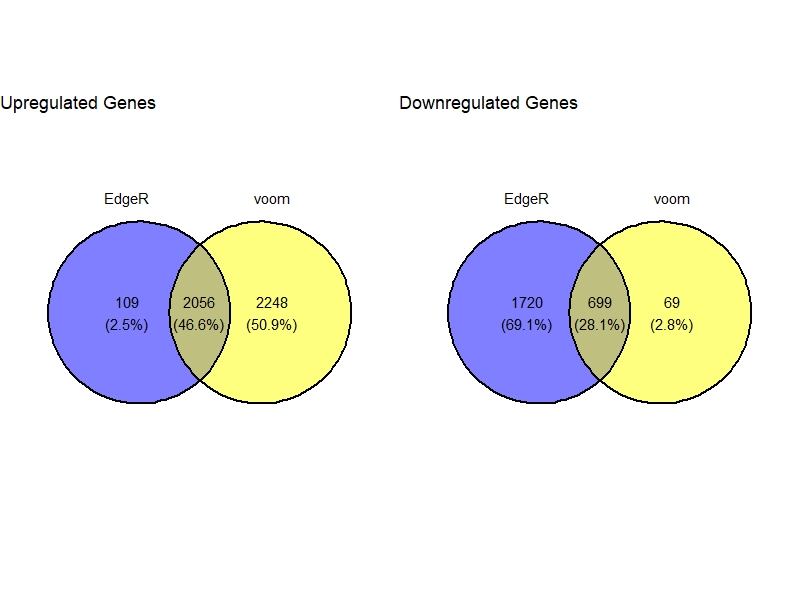
Plots
Prepare the data.
data("DEG")
DE.list<-list("edger" =dge_edger, "edgerql" = dge_edgerql, "deseq2" = dge_deseq2, "voom" = dge_voom )
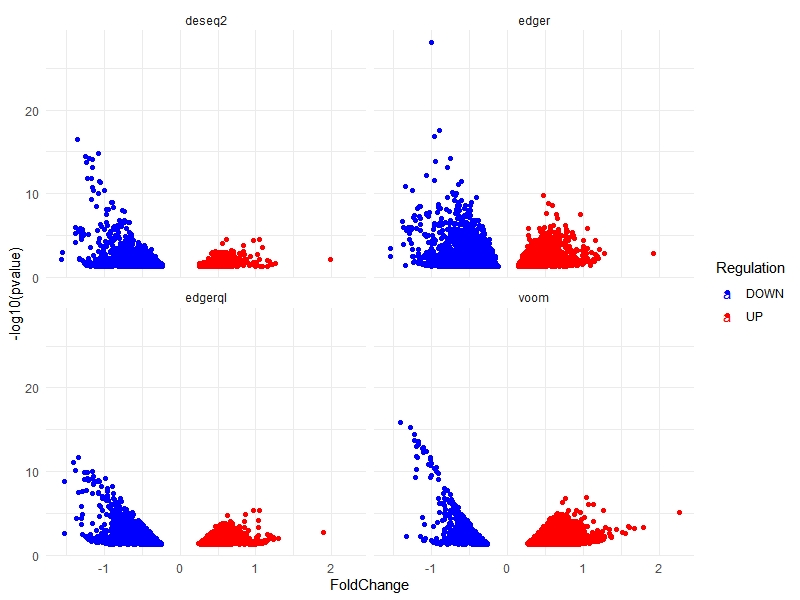
Volcano plots
multiVolcano(DE.list, FoldChange = 1.4, DE.Only = FALSE , show.genes = TRUE )
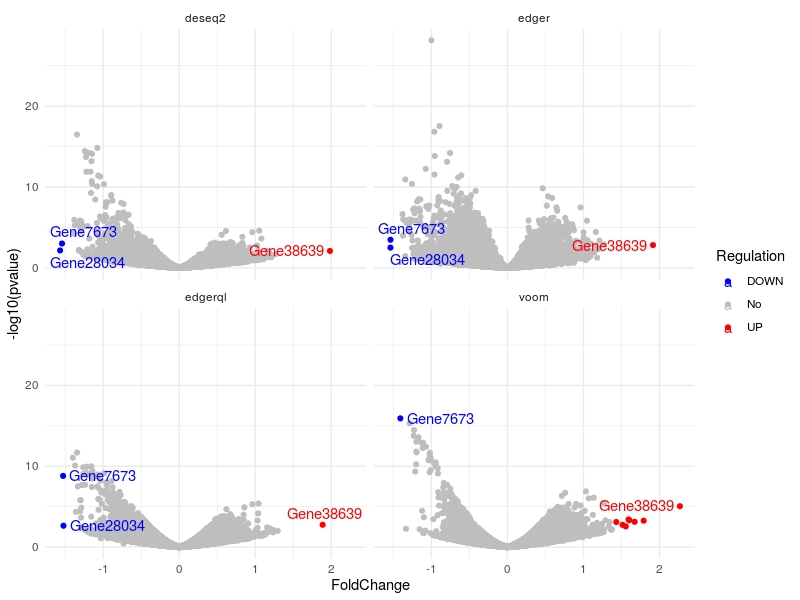
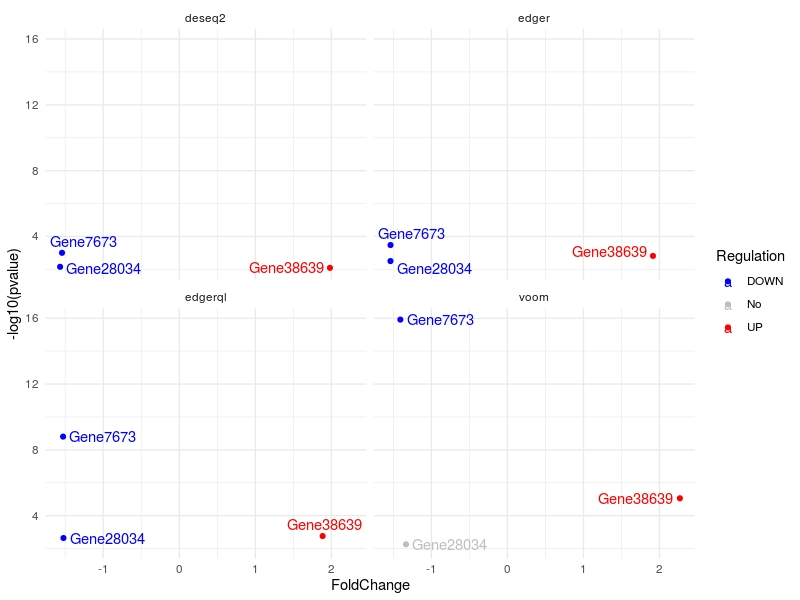
For user defined gene list
genes.list<-c("Gene7673", "Gene28034", "Gene38639")
multiVolcano(DE.list, FoldChange = 1.4, DE.Only = FALSE , genes = genes.list , show.genes = TRUE)
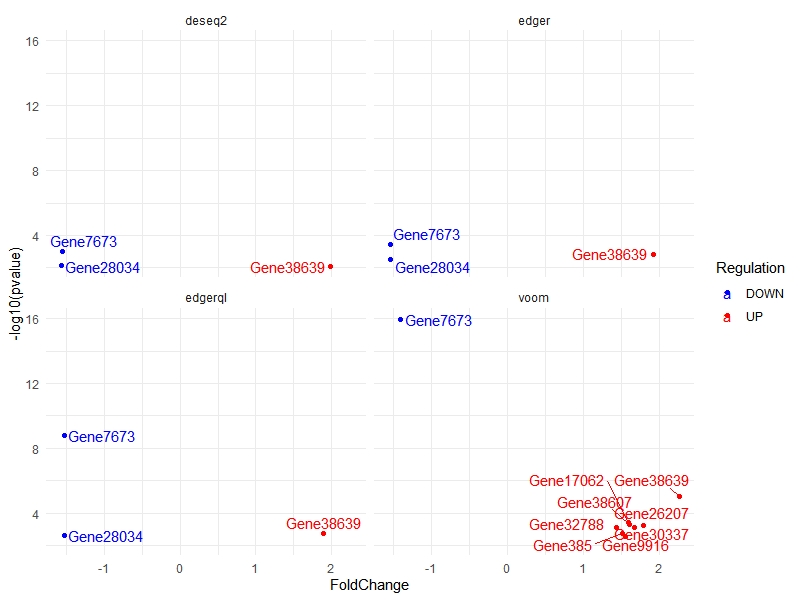
For showing Differentially expressed genes. This command will show only a few of the outlier genes)
multiVolcano(DE.list, show.genes = FALSE)
For showing Differentially expressed genes (marking show.genes as false for more asthetic plot)
multiVolcano(DE.list, FoldChange = 1.4 , show.genes = FALSE)
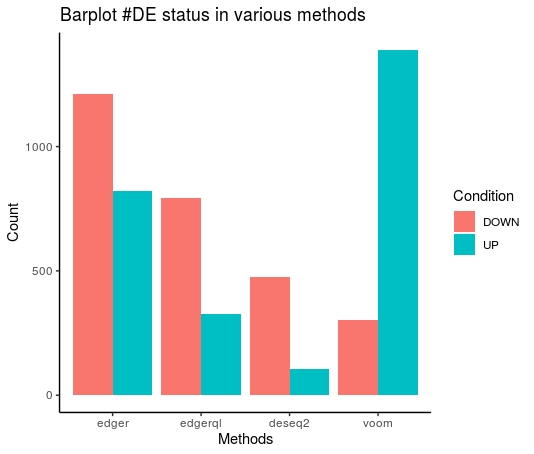
Bar plot
TabulateStats(DE=DE.list , FoldChange=0 , cutoff =0.01 )
Tabular Result
| |edger|edgerql|voom|
|:----:|:----:|:------:|:-----:|
|DOWN|1210|794|302|
|No|33947|34858|34287|
|UP| 821|326|1389|
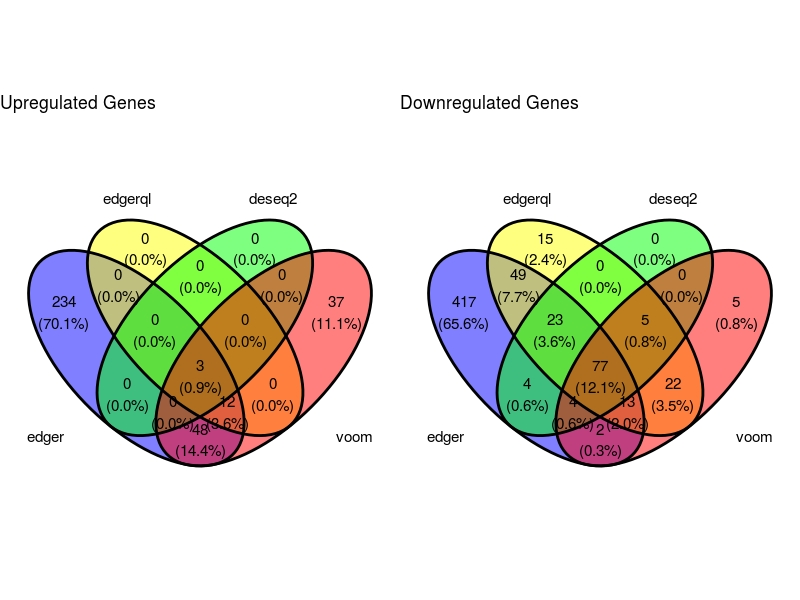
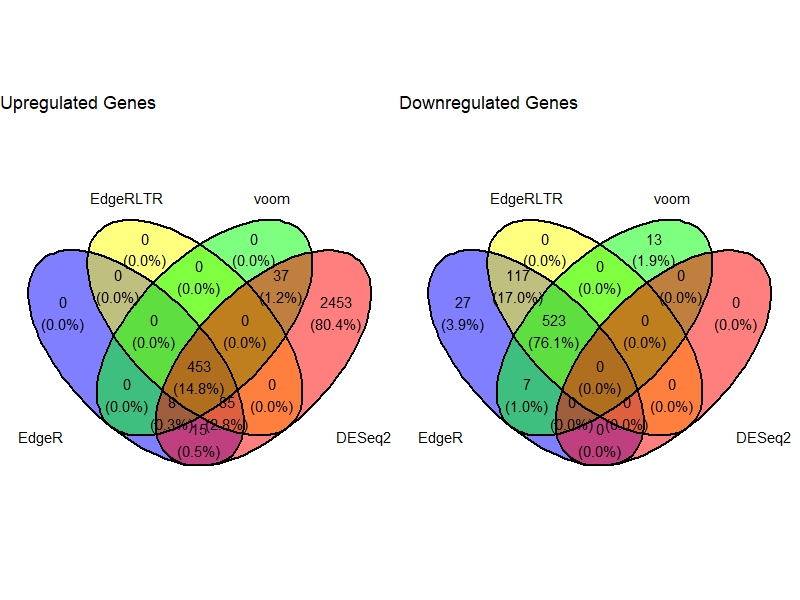
Venn Diagram
VennDig(DE=DE.list , FoldChange=1.5 , cutoff =0.01 , type.sig ="FDR")
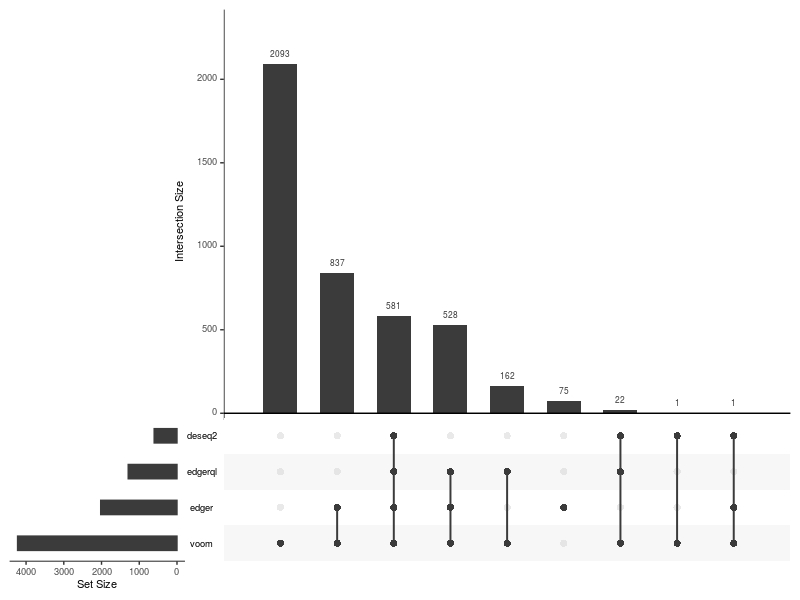
Upset plot
UpSetPlot(DE=DE.list , FoldChange=0 , cutoff =0.01 , type.sig = "p")
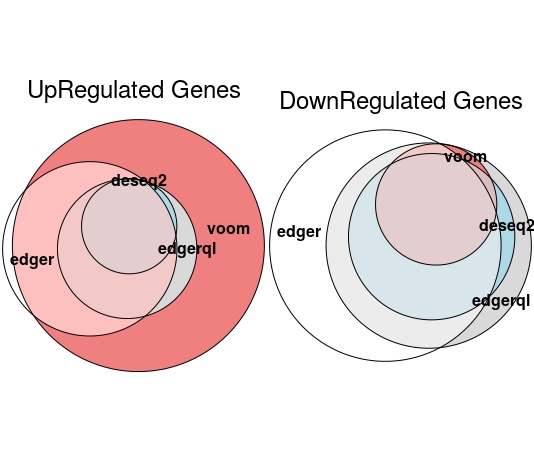
Prapotional Venn Diagram
EulerrPlot(DE=DE.list , FoldChange=0 , cutoff =0.05 , type.sig ="p")
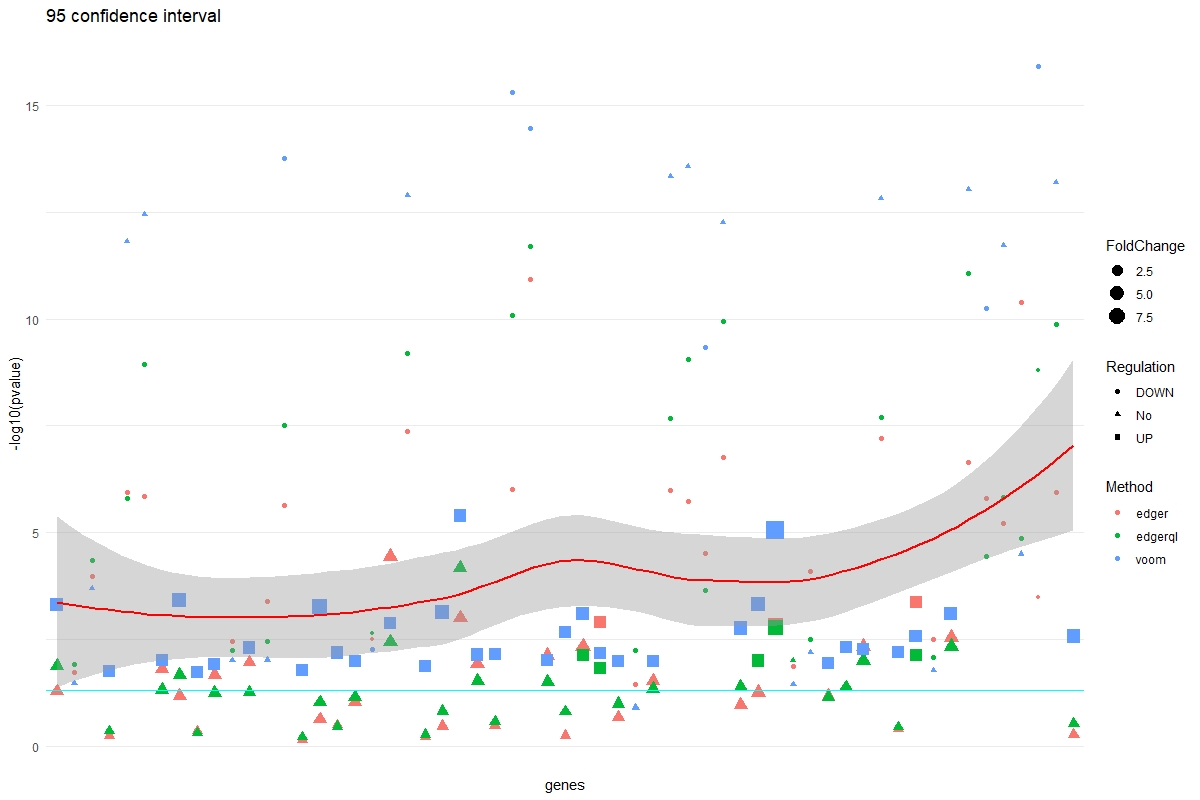
CI interval plot and data frame
DGE.CI(DE=DE.list , FoldChange=1.2 , cutoff =0.05 , type.sig ="p")
| |edger FoldChange|edgerql FoldChange|voom FoldChange|Min|Max|edger pvalue|edgerql pvalue|voom pvalue|Min|Max|
|:--:|:--:|:--:|:--:|:--:|:--:|:--:|:--:|:--:|:--:|:--:|
|Gene11876|0.253268010646449|0.25665960045484|0.333519105983995|0.253268010646449|0.333519105983995|0.00966910768271969|0.0138679244791296|0.0390346542589136|0.00966910768271969|0.0390346542589136|
|Gene15604|0.27180343736292|0.271787631013647|0.302097146830169|0.271787631013647|0.302097146830169|0.000348901761561591|0.00103259777034587|3.97342702281636e-09|3.97342702281636e-09|0.00103259777034587|
|Gene15888|0.30094351744471|0.300302928429722|0.332617475249204|0.300302928429722|0.332617475249204|0.000405266030230859|2.99397057448929e-06|1.16326308848636e-09|1.16326308848636e-09|0.000405266030230859|
|Gene22288|0.263474804206977|0.263400660850057|0.2916799978793|0.263400660850057|0.2916799978793|0.0005861510066759|4.17234374228856e-05|1.5530006115196e-10|1.5530006115196e-10|0.0005861510066759|
|Gene29009|0.294253032199853|0.293993599013282|0.330000812105133|0.293993599013282|0.330000812105133|2.62505647540443e-05|2.81215059098373e-06|5.0579118548581e-10|5.0579118548581e-10|2.62505647540443e-05|
|Gene30812|3.0011502354894|2.82286115292464|3.54654652335402|2.82286115292464|3.54654652335402|0.0443053766662665|0.0192060783610423|0.00229564569695183|0.00229564569695183|0.0443053766662665|
|Gene32136|0.253951391140971|0.253847733944933|0.276660037325252|0.253847733944933|0.276660037325252|0.000311904095164778|8.10948071015179e-07|8.89107509409887e-12|8.89107509409887e-12|0.000311904095164778|
|Gene3219|0.26132542426818|0.260852100787794|0.292821559694857|0.260852100787794|0.292821559694857|3.9108014179455e-08|7.25440228196772e-08|4.04635632813972e-11|4.04635632813972e-11|7.25440228196772e-08|
|Gene3636|0.276773935358202|0.276658009021903|0.30742464752486|0.276658009021903|0.30742464752486|0.000320786232115754|3.12890638531982e-05|2.80586403681253e-10|2.80586403681253e-10|0.000320786232115754|
|Gene36720|0.282886374057711|0.283071871234626|0.310527439322474|0.282886374057711|0.310527439322474|0.000489633506875775|2.9117905222642e-06|1.90836815806636e-10|1.90836815806636e-10|0.000489633506875775|
|Gene37809|0.274775812479113|0.274781151367946|0.297749013272049|0.274775812479113|0.297749013272049|0.00409971202637718|0.0403344717227362|5.83264594328474e-07|5.83264594328474e-07|0.0403344717227362|
|Gene38199|0.295370543964787|0.296061444016233|0.328422674982562|0.295370543964787|0.328422674982562|8.2482427048141e-05|8.10948071015179e-07|1.52215838845913e-09|1.52215838845913e-09|8.2482427048141e-05|
|Gene38639|6.7835229404579|6.58741660386054|9.6638046407115|6.58741660386054|9.6638046407115|0.0573934369490823|0.136024039700254|0.00415995911576501|0.00415995911576501|0.136024039700254|
|Gene38937|0.283452115852866|0.27553856211236|0.315105821098576|0.27553856211236|0.315105821098576|0.00825053138204574|0.195476227936559|0.181147657675811|0.00825053138204574|0.195476227936559|
|Gene40770|0.289698705765732|0.289780204987156|0.325317239975445|0.289698705765732|0.325317239975445|3.4623122963608e-05|3.08747716025276e-05|5.52436063074383e-10|5.52436063074383e-10|3.4623122963608e-05|
|Gene6678|0.251819899040539|0.246647238865664|0.301947985426532|0.246647238865664|0.301947985426532|0.00010352524415427|1.56612785739337e-07|4.16666249706773e-10|4.16666249706773e-10|0.00010352524415427|
|Gene6710|0.271459164291596|0.27145692542583|0.301120050678203|0.27145692542583|0.301120050678203|0.000430650590640714|0.0118339982149324|9.04425211253826e-08|9.04425211253826e-08|0.0118339982149324|
|Gene720|0.273484715998214|0.273434784275442|0.302884849886368|0.273434784275442|0.302884849886368|0.00122751324077398|0.00102621047894686|4.53220294233197e-09|4.53220294233197e-09|0.00122751324077398|
|Gene7339|0.284743280003845|0.275612814902474|0.32548462536702|0.275612814902474|0.32548462536702|1.22959638267496e-07|0.00588064870290072|0.0108920996649206|1.22959638267496e-07|0.0108920996649206|
|Gene7673|0.21499523132271|0.216980068912695|0.244685085090055|0.21499523132271|0.244685085090055|0.0218327855690298|3.60045900816876e-06|4.35821145336272e-12|4.35821145336272e-12|0.0218327855690298|
|Gene9230|0.282854723987658|0.282620482988113|0.310124743179966|0.282620482988113|0.310124743179966|0.000351248577561045|8.10948071015179e-07|3.29044695217997e-10|3.29044695217997e-10|0.000351248577561045|
Golub
# Install the required packages
BiocManager::install("multtest")
library(multtest)
# load the golub dataset
data(golub)
ds<-data.frame("trt" = golub.cl)
colnames(golub) <-paste0("Name",1:length(golub.cl))
row.names(ds) <- colnames(golub)
ds$trt <- paste0("C",ds$trt)
xx<-as.data.frame(round(exp(golub)*1000))
DGE.list <- DGE(count.table = xx , design.matrix = ds )
VennDig(DGE.list)
DGE.CI(DE=DE.list , FoldChange=1.2 , cutoff =0.05 , type.sig ="p")
afitz-gmu/DGE-Analysis documentation built on April 3, 2021, 2:39 a.m.
DGE-Analysis
Index
Install
Install the required packages and link to the correct github repository.
library(remotes)
remotes::install_github(repo="afitz-gmu/DGE-Analysis", build_opts = c("--no-resave-data", "--no-manual"))
library(GenomicVisualization)
DGE
Basic function to perfrom Differential Gene Expression by EdgeR, DESeq2 and Voom.
data("count.table")
design <- data.frame("trt" = colnames(count.table))
rownames(design) <- design$trt
design$trt <- as.integer(grepl("T",design[,1]))
DGE.list <- DGE(count.table = count.table , design.matrix = design , method=c("EdgeR","voom") )
VennDig(DGE.list)

Plots
Prepare the data.
data("DEG")
DE.list<-list("edger" =dge_edger, "edgerql" = dge_edgerql, "deseq2" = dge_deseq2, "voom" = dge_voom )
Volcano plots
multiVolcano(DE.list, FoldChange = 1.4, DE.Only = FALSE , show.genes = TRUE )

For user defined gene list
genes.list<-c("Gene7673", "Gene28034", "Gene38639")
multiVolcano(DE.list, FoldChange = 1.4, DE.Only = FALSE , genes = genes.list , show.genes = TRUE)

For showing Differentially expressed genes. This command will show only a few of the outlier genes)
multiVolcano(DE.list, show.genes = FALSE)

For showing Differentially expressed genes (marking show.genes as false for more asthetic plot)
multiVolcano(DE.list, FoldChange = 1.4 , show.genes = FALSE)

Bar plot
TabulateStats(DE=DE.list , FoldChange=0 , cutoff =0.01 )

Tabular Result
| |edger|edgerql|voom| |:----:|:----:|:------:|:-----:| |DOWN|1210|794|302| |No|33947|34858|34287| |UP| 821|326|1389|
Venn Diagram
VennDig(DE=DE.list , FoldChange=1.5 , cutoff =0.01 , type.sig ="FDR")

Upset plot
UpSetPlot(DE=DE.list , FoldChange=0 , cutoff =0.01 , type.sig = "p")

Prapotional Venn Diagram
EulerrPlot(DE=DE.list , FoldChange=0 , cutoff =0.05 , type.sig ="p")

CI interval plot and data frame
DGE.CI(DE=DE.list , FoldChange=1.2 , cutoff =0.05 , type.sig ="p")

| |edger FoldChange|edgerql FoldChange|voom FoldChange|Min|Max|edger pvalue|edgerql pvalue|voom pvalue|Min|Max| |:--:|:--:|:--:|:--:|:--:|:--:|:--:|:--:|:--:|:--:|:--:| |Gene11876|0.253268010646449|0.25665960045484|0.333519105983995|0.253268010646449|0.333519105983995|0.00966910768271969|0.0138679244791296|0.0390346542589136|0.00966910768271969|0.0390346542589136| |Gene15604|0.27180343736292|0.271787631013647|0.302097146830169|0.271787631013647|0.302097146830169|0.000348901761561591|0.00103259777034587|3.97342702281636e-09|3.97342702281636e-09|0.00103259777034587| |Gene15888|0.30094351744471|0.300302928429722|0.332617475249204|0.300302928429722|0.332617475249204|0.000405266030230859|2.99397057448929e-06|1.16326308848636e-09|1.16326308848636e-09|0.000405266030230859| |Gene22288|0.263474804206977|0.263400660850057|0.2916799978793|0.263400660850057|0.2916799978793|0.0005861510066759|4.17234374228856e-05|1.5530006115196e-10|1.5530006115196e-10|0.0005861510066759| |Gene29009|0.294253032199853|0.293993599013282|0.330000812105133|0.293993599013282|0.330000812105133|2.62505647540443e-05|2.81215059098373e-06|5.0579118548581e-10|5.0579118548581e-10|2.62505647540443e-05| |Gene30812|3.0011502354894|2.82286115292464|3.54654652335402|2.82286115292464|3.54654652335402|0.0443053766662665|0.0192060783610423|0.00229564569695183|0.00229564569695183|0.0443053766662665| |Gene32136|0.253951391140971|0.253847733944933|0.276660037325252|0.253847733944933|0.276660037325252|0.000311904095164778|8.10948071015179e-07|8.89107509409887e-12|8.89107509409887e-12|0.000311904095164778| |Gene3219|0.26132542426818|0.260852100787794|0.292821559694857|0.260852100787794|0.292821559694857|3.9108014179455e-08|7.25440228196772e-08|4.04635632813972e-11|4.04635632813972e-11|7.25440228196772e-08| |Gene3636|0.276773935358202|0.276658009021903|0.30742464752486|0.276658009021903|0.30742464752486|0.000320786232115754|3.12890638531982e-05|2.80586403681253e-10|2.80586403681253e-10|0.000320786232115754| |Gene36720|0.282886374057711|0.283071871234626|0.310527439322474|0.282886374057711|0.310527439322474|0.000489633506875775|2.9117905222642e-06|1.90836815806636e-10|1.90836815806636e-10|0.000489633506875775| |Gene37809|0.274775812479113|0.274781151367946|0.297749013272049|0.274775812479113|0.297749013272049|0.00409971202637718|0.0403344717227362|5.83264594328474e-07|5.83264594328474e-07|0.0403344717227362| |Gene38199|0.295370543964787|0.296061444016233|0.328422674982562|0.295370543964787|0.328422674982562|8.2482427048141e-05|8.10948071015179e-07|1.52215838845913e-09|1.52215838845913e-09|8.2482427048141e-05| |Gene38639|6.7835229404579|6.58741660386054|9.6638046407115|6.58741660386054|9.6638046407115|0.0573934369490823|0.136024039700254|0.00415995911576501|0.00415995911576501|0.136024039700254| |Gene38937|0.283452115852866|0.27553856211236|0.315105821098576|0.27553856211236|0.315105821098576|0.00825053138204574|0.195476227936559|0.181147657675811|0.00825053138204574|0.195476227936559| |Gene40770|0.289698705765732|0.289780204987156|0.325317239975445|0.289698705765732|0.325317239975445|3.4623122963608e-05|3.08747716025276e-05|5.52436063074383e-10|5.52436063074383e-10|3.4623122963608e-05| |Gene6678|0.251819899040539|0.246647238865664|0.301947985426532|0.246647238865664|0.301947985426532|0.00010352524415427|1.56612785739337e-07|4.16666249706773e-10|4.16666249706773e-10|0.00010352524415427| |Gene6710|0.271459164291596|0.27145692542583|0.301120050678203|0.27145692542583|0.301120050678203|0.000430650590640714|0.0118339982149324|9.04425211253826e-08|9.04425211253826e-08|0.0118339982149324| |Gene720|0.273484715998214|0.273434784275442|0.302884849886368|0.273434784275442|0.302884849886368|0.00122751324077398|0.00102621047894686|4.53220294233197e-09|4.53220294233197e-09|0.00122751324077398| |Gene7339|0.284743280003845|0.275612814902474|0.32548462536702|0.275612814902474|0.32548462536702|1.22959638267496e-07|0.00588064870290072|0.0108920996649206|1.22959638267496e-07|0.0108920996649206| |Gene7673|0.21499523132271|0.216980068912695|0.244685085090055|0.21499523132271|0.244685085090055|0.0218327855690298|3.60045900816876e-06|4.35821145336272e-12|4.35821145336272e-12|0.0218327855690298| |Gene9230|0.282854723987658|0.282620482988113|0.310124743179966|0.282620482988113|0.310124743179966|0.000351248577561045|8.10948071015179e-07|3.29044695217997e-10|3.29044695217997e-10|0.000351248577561045|
Golub
# Install the required packages
BiocManager::install("multtest")
library(multtest)
# load the golub dataset
data(golub)
ds<-data.frame("trt" = golub.cl)
colnames(golub) <-paste0("Name",1:length(golub.cl))
row.names(ds) <- colnames(golub)
ds$trt <- paste0("C",ds$trt)
xx<-as.data.frame(round(exp(golub)*1000))
DGE.list <- DGE(count.table = xx , design.matrix = ds )
VennDig(DGE.list)

DGE.CI(DE=DE.list , FoldChange=1.2 , cutoff =0.05 , type.sig ="p")
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
