README.md
In js2264/periodicDNA: Set of tools to identify periodic occurrences of k-mers in DNA sequences
[ ](https://f1000research.com/articles/10-141)
[
](https://f1000research.com/articles/10-141)
[ ](https://doi.org/10.1101/gr.265934.120)
](https://doi.org/10.1101/gr.265934.120)






periodicDNA 
Introduction
This R package helps the user identify k-mers (e.g. di- or
tri-nucleotides) present periodically in a set of genomic loci (typically
regulatory elements). It is not aimed at identifying motifs separated by a
conserved distance; for this type of analysis, please visit
MEME website.
Installation
periodicDNA is available in Bioconductor. To install the current release use:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("periodicDNA")
For advanced users, the most recent periodicDNA can be installed
from Github as follows (might be buggy):
install.packages("devtools")
devtools::install_github("js2264/periodicDNA")
library(periodicDNA)
Citation
If you are using periodicDNA in your research, please cite:
periodicDNA: an R/Bioconductor package to investigate k-mer periodicity in DNA
J. Serizay & J. Ahringer
F1000Research, 2021
Distinctive regulatory architectures of germline-active and somatic genes in C. elegans
J. Serizay, Y. Dong, J. Jänes, M. Chesney, C. Cerrato & J. Ahringer
Genome Research, 2020
How to use periodicDNA
periodicDNA includes a vignette where its usage is
illustrated. To access the vignette, please use:
vignette('periodicDNA')
Main functions
The two main user-level functions of periodicDNA are getPeriodicity() and
getPeriodicityTrack().
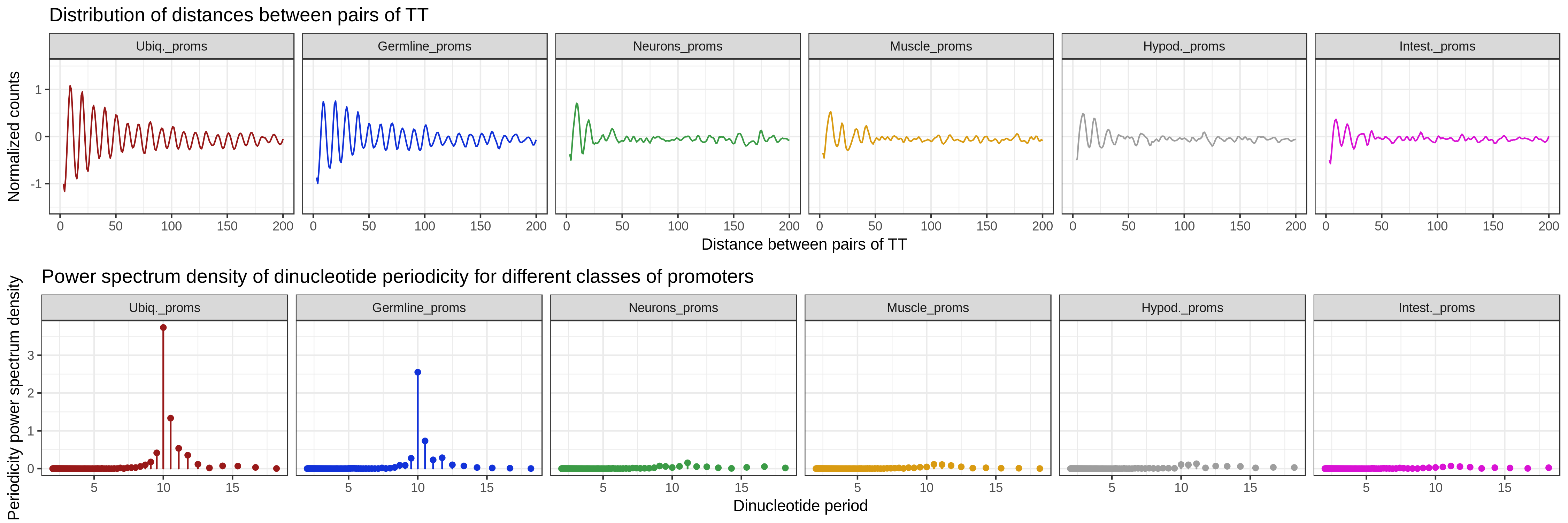
getPeriodicity() is used to compute the power spectral density
(PSD) of a chosen k-mer (i.e. TT) in a set of sequences. The PSD
score at a given period indicates the strength of the k-mer at
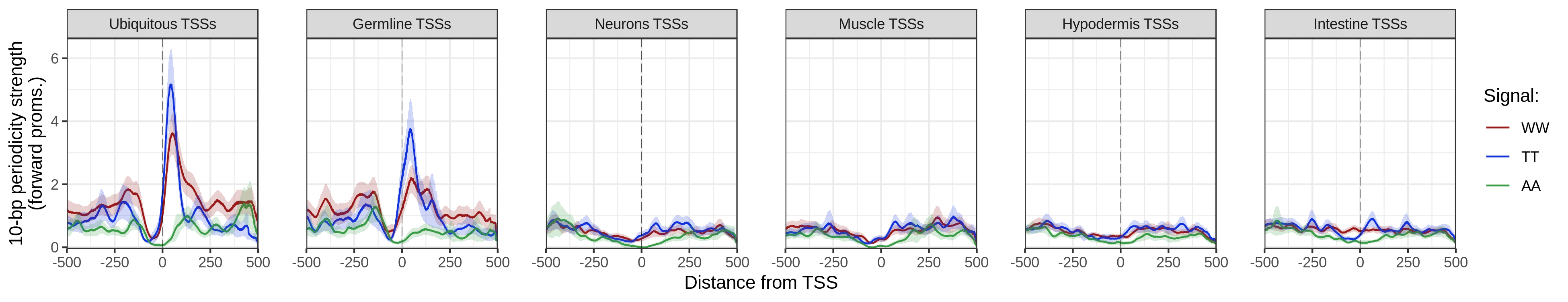
this period. getPeriodicityTrack() can be used to generate linear tracks representing
the periodicity strength of a given k-mer at a chosen period, over genomic
loci of interest.
getPeriodicity() function
data(ce11_TSSs)
PSDs <- getPeriodicity(
ce11_TSSs[['Ubiq.']],
genome = 'BSgenome.Celegans.UCSC.ce11',
motif = 'TT',
BPPARAM = MulticoreParam(12),
n_shuffling = 100
)
plotPeriodicityResults(PSDs)
getPeriodicityTrack() function
data(ce11_proms)
WW_10bp <- getPeriodicityTrack(
genome = 'BSgenome.Celegans.UCSC.ce11',
granges = ce11_proms,
motif = 'WW',
period = 10,
bw_file = 'WW-10-bp-periodicity_over-proms.bw',
BPPARAM = MulticoreParam(12)
)
Warning: It is recommended to run this command across many processors
using BiocParallel. This command typically takes one day to produce
a periodicity track over 15,000 GRanges of 150 bp (with default parameters)
using BPPARAM = MulticoreParam(12).
It is highly recommended to run this command in a new screen session.
Contributions
Code contributions, bug reports, fixes and feature requests are most welcome.
Please make any pull requests against the master branch at
https://github.com/js2264/periodicityDNA
and file issues at https://github.com/js2264/periodicityDNA/issues
License
periodicDNA is licensed under the GPL-3 license.
js2264/periodicDNA documentation built on Nov. 3, 2022, 10:47 p.m.
[





periodicDNA 


Introduction
This R package helps the user identify k-mers (e.g. di- or tri-nucleotides) present periodically in a set of genomic loci (typically regulatory elements). It is not aimed at identifying motifs separated by a conserved distance; for this type of analysis, please visit MEME website.
Installation
periodicDNA is available in Bioconductor. To install the current release use:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("periodicDNA")
For advanced users, the most recent periodicDNA can be installed from Github as follows (might be buggy):
install.packages("devtools")
devtools::install_github("js2264/periodicDNA")
library(periodicDNA)
Citation
If you are using periodicDNA in your research, please cite:
periodicDNA: an R/Bioconductor package to investigate k-mer periodicity in DNA J. Serizay & J. Ahringer F1000Research, 2021
Distinctive regulatory architectures of germline-active and somatic genes in C. elegans J. Serizay, Y. Dong, J. Jänes, M. Chesney, C. Cerrato & J. Ahringer Genome Research, 2020
How to use periodicDNA
periodicDNA includes a vignette where its usage is illustrated. To access the vignette, please use:
vignette('periodicDNA')
Main functions
The two main user-level functions of periodicDNA are getPeriodicity() and
getPeriodicityTrack().
getPeriodicity()is used to compute the power spectral density (PSD) of a chosen k-mer (i.e.TT) in a set of sequences. The PSD score at a given period indicates the strength of the k-mer at this period.getPeriodicityTrack()can be used to generate linear tracks representing the periodicity strength of a given k-mer at a chosen period, over genomic loci of interest.
getPeriodicity() function
data(ce11_TSSs)
PSDs <- getPeriodicity(
ce11_TSSs[['Ubiq.']],
genome = 'BSgenome.Celegans.UCSC.ce11',
motif = 'TT',
BPPARAM = MulticoreParam(12),
n_shuffling = 100
)
plotPeriodicityResults(PSDs)
getPeriodicityTrack() function
data(ce11_proms)
WW_10bp <- getPeriodicityTrack(
genome = 'BSgenome.Celegans.UCSC.ce11',
granges = ce11_proms,
motif = 'WW',
period = 10,
bw_file = 'WW-10-bp-periodicity_over-proms.bw',
BPPARAM = MulticoreParam(12)
)
Warning: It is recommended to run this command across many processors
using BiocParallel. This command typically takes one day to produce
a periodicity track over 15,000 GRanges of 150 bp (with default parameters)
using BPPARAM = MulticoreParam(12).
It is highly recommended to run this command in a new screen session.
Contributions
Code contributions, bug reports, fixes and feature requests are most welcome. Please make any pull requests against the master branch at https://github.com/js2264/periodicityDNA and file issues at https://github.com/js2264/periodicityDNA/issues
License
periodicDNA is licensed under the GPL-3 license.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
