In MonkeyLB/hicrep: Measuring the reproducibility of Hi-C data
Introduction
Hi-C data analysis and interpretation are still in their early stages.
In particular, there has been a lack of sound statistical metric to
evaluate the quality of Hi-C data. When biological replicates are not
available, investigators often rely on eithervisual inspection of Hi-C
interaction heatmap or examining the ratio of long-range interaction
read pairs over the total sequenced reads, neither of which are supported
by robust statistics. When two or more biological replicates are available,
it is a common practice to compute either Pearson or Spearman correlation
coefficients between the two Hi-C data matrices and use them as a metric
for quality control. However, these kind of over-simplified approaches are
problematic and may lead to wrong conclusions, because they do not take
into consideration of the unique characteristics of Hi-C data, such as
distance-dependence and domain structures. As a result, two un-related
biological samples can have a strong Pearson correlation coefficient, while
two visually similar replicates can have poor Spearman correlation coefficient.
It is also not uncommon to observe higher Pearson and Spearman correlations
between unrelated samples than those between real biological replicates.
we develop a novel framework, hicrep, for assessing the reproducibility of
Hi-C data. It first minimizes the effect of noise and biases by smoothing
Hi-C matrix, and then addresses the distance-dependence effect by stratifying
Hi-C data according to their genomic distance. We further adopt a
stratum-adjusted correlation coefficient (SCC) as the measurement of Hi-C data
reproducibility. The value of SCC ranges from -1 to 1, and it can be used to
compare the degrees of differences in reproducibility. Our framework can also
infer confidence intervals for SCC, and further estimate the statistical
significance of the difference in reproducibility measurement for different
data sets.
In this Vignette, we explain the method rationale, and provide guidance to
use the functions of hicrep to assess the reproducibility for Hi-C
intrachromosome replicates.
Citation
Cite our paper:
HiCRep: assessing the reproducibility of Hi-C data using a
stratum-adjusted correlation coefficient. Tao Yang, Feipeng Zhang, Galip
Gurkan Yardimci, Fan Song, Ross C Hardison, William Stafford Noble,
Feng Yue, Qunhua Li. Genome Research 2017. doi: 10.1101/gr.220640.117.
Installation
Download the source package (hicrep_xxx.tar.gz) from Github.
Or install it from Bioconductor:
## try http:// if https:// URLs are not supported
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("hicrep")
Rationale of method
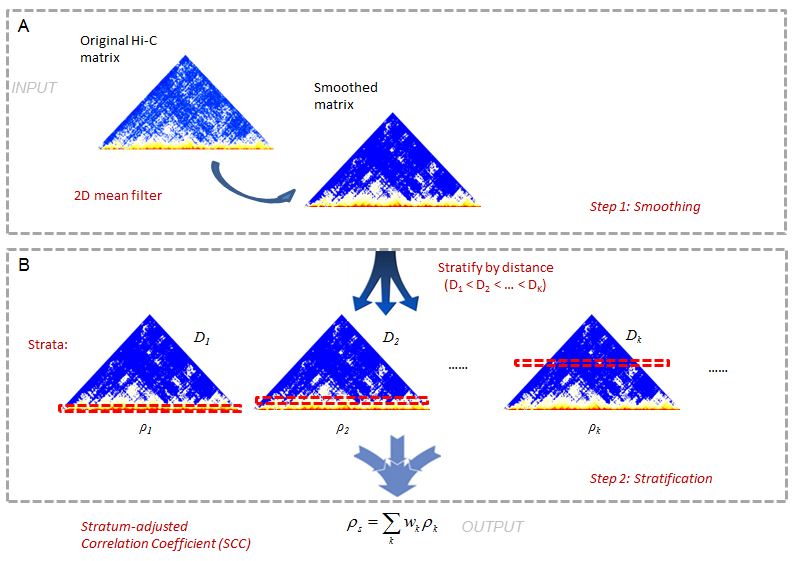
This is a 2-step method (Figure1). In Hi-C data it is often difficult to
achieve sufficient coverage. When samples are not sufficiently sequenced,
the local variation introduced by under-sampling can make it difficult to
capture large domain structures. To reduce local variation, we first smooth
the contact map before assessing reproducibility. Although a smoothing filter
will reduce the individual spatial resolution, it can improve the contiguity
of the regions with elevated interaction, consequently enhancing the domain
structures. We use a 2D moving window average filter to smooth the Hi-C
contact map. This choice is made for the simplicity and fast computation of
mean filter, and the rectangular shape of Hi-C compartments.
In the second step, we stratify the Hi-C reads by the distance of contacting
loci, calculate the Pearson correlations within each stratum, and then
summarize the stratum-specific correlation coefficients into an aggregated
statistic. We name it as Stratum-adjusted Correlation Coefficient (SCC).
For the methodology details, please refer to our paper on Genome Research.
knitr::opts_knit$set(progress = TRUE, verbose = TRUE)
library(hicrep)
data("HiCR1")
data("HiCR2")
The format of input and Pre-processing
The input are two Hi-C matrices to be compared. The Hi-C matrices
should have the dimension $N\times N$.
dim(HiCR1)
HiCR1[1:10,1:10]
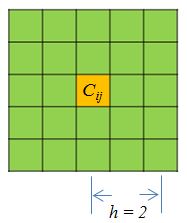
The function get.scc will first smooth the HiC matrix, with given
neighborhood size parameter $h$, and filter the bins that have
zero counts in both replicates. The arguments includes the two matrices,
the resolution of matrices, smoothing parameter, and the lower bound and
upper bound of interaction distance considered. The resolution is simply
the bin size. Smoothing parameter decides the neighborhood size of
smoothing. Below (Figure 2) is a representation of smoothing neighborhood
for a point $C_{ij}$:
Calculate Stratum-adjusted Correlation Coefficient (SCC)
An example to calculate SCC for a matrix of 1Mb resolution. Smoothing
parameter $h$ is set to 2. The lower bound of distance considered is 0
(diagnal), and the upper bound is 5Mb.
scc.out = get.scc(HiCR1, HiCR2, 1000000, 2, 0, 5000000)
#SCC score
scc.out$scc
#Standard deviation of SCC
scc.out$std
The output is a list of results including stratum specific Pearson
correlations, weight coefficient, SCC and the asymptotic standard
deviation of SCC. The last two numbers are the ones we needed in most
of the situations.
Smooth the Hi-C matrix with 2D mean filter
The function fast.mean.filter() is a very fast algorithm that applies
2D mean filter to squred matrices such as Hi-C contact mapes. The output
is a smoothed matrix that has the same size with the original matrix.
Here is an example to smooth the matrix with parameter $h = 2$:
smd_mat = fast.mean.filter(HiCR1, 2)
Select the optimal smoothing parameter
To select $h$ objectively, we develop a heuristic procedure to search
for the optimal smoothing parameter. Our procedure is designed based
on the observation that the correlation between contact maps of replicate
samples first increases with the level of smoothness and plateaus when
sufficient smoothness is reached.To proceed, we use a pair of reasonably
deeply sequenced interaction map as the training data. We randomly
sampled 10% of the data, then compute SCC for the sampled dataeach
fraction at a series of smoothing parameters in the ascending order. We
choose the smallest $h$ at which the increment of the average
reproducibility score is less than 0.01. This procedure is repeated ten
times, and the mode among the ten $h$’s is picked.
h_hat <- htrain(HiCR1, HiCR2, 1000000, lbr = 0, ubr = 5000000, range = 0:2)
h_hat
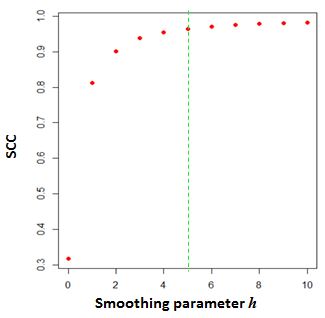
The above graph shows the change of SCC as the $h$ increases from 0 to 10
for a 40Kb resolution matrix. The parameter $h = 5$ is selected as the
optimal smoothing neighborhood size.
Important note:
The smoothing parameter selection could be confounded by the sequencing
depth. Insufficient sequencing depth data might lead to inflated
smoothing neighborhood size. To compare SCC between pairs of
replicates that has the same resolution, one shall use the same
smoothing parameter.
Train the smoothing parameter could be time-consuming. It is not
suggested to train $h$ every time when calculating SCC. For a giving
resolution, one could use a deeply sequenced biological replicates
to train $h$ (i.e., > 300 million total nubmer of reads for whole
chromosome), and use the trained $h$ for other same resolution data.
Here we provide some trained $h$ trained based on two replicates
of hESC cells from Dixon et al 2015 (GEO accession: GSE52457):
Resolution ($h$):
10K (20), 25K (10), 40k(5), 100k(3), 500k(1 or 2), 1M(0 or 1).
Equalize the total number of reads
In previous section, we mention that sequencing depth could be a confounding
effect. If the total numbers of reads are very different between the two
replicates, it's suggested that one should down-sample the higher sequencing
depth to make it equal to the lower one. The best way to do it is to use the
bam files to do the sub-sampling randomly. In case you only have the matrix
file available, we made a function depth.adj() to do down-sampling from
matrix files.
#check total number of reads before adjustment
sum(HiCR1)
# sub-sample 200000 total reads
DS_HiCR1 <- depth.adj(HiCR1, 200000)
#check total number of reads after adjustment
sum(DS_HiCR1)
sessionInfo()
#check total number of reads before adjustment
sum(HiCR1)
# sub-sample 200000 total reads
DS_HiCR1 <- depth.adj(HiCR1, 200000)
#check total number of reads after adjustment
sum(DS_HiCR1)
Computation efficiency
Given a pair of contact maps of human chromosome 1 with bin-size equal to
40kb, it takes 27 seconds on a laptop with 2.6GHz Intel Core i7-6600U and
16Gb of RAM.
MonkeyLB/hicrep documentation built on Dec. 15, 2020, 12:47 a.m.
Introduction
Hi-C data analysis and interpretation are still in their early stages. In particular, there has been a lack of sound statistical metric to evaluate the quality of Hi-C data. When biological replicates are not available, investigators often rely on eithervisual inspection of Hi-C interaction heatmap or examining the ratio of long-range interaction read pairs over the total sequenced reads, neither of which are supported by robust statistics. When two or more biological replicates are available, it is a common practice to compute either Pearson or Spearman correlation coefficients between the two Hi-C data matrices and use them as a metric for quality control. However, these kind of over-simplified approaches are problematic and may lead to wrong conclusions, because they do not take into consideration of the unique characteristics of Hi-C data, such as distance-dependence and domain structures. As a result, two un-related biological samples can have a strong Pearson correlation coefficient, while two visually similar replicates can have poor Spearman correlation coefficient. It is also not uncommon to observe higher Pearson and Spearman correlations between unrelated samples than those between real biological replicates.
we develop a novel framework, hicrep, for assessing the reproducibility of
Hi-C data. It first minimizes the effect of noise and biases by smoothing
Hi-C matrix, and then addresses the distance-dependence effect by stratifying
Hi-C data according to their genomic distance. We further adopt a
stratum-adjusted correlation coefficient (SCC) as the measurement of Hi-C data
reproducibility. The value of SCC ranges from -1 to 1, and it can be used to
compare the degrees of differences in reproducibility. Our framework can also
infer confidence intervals for SCC, and further estimate the statistical
significance of the difference in reproducibility measurement for different
data sets.
In this Vignette, we explain the method rationale, and provide guidance to
use the functions of hicrep to assess the reproducibility for Hi-C
intrachromosome replicates.
Citation
Cite our paper:
HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Tao Yang, Feipeng Zhang, Galip Gurkan Yardimci, Fan Song, Ross C Hardison, William Stafford Noble, Feng Yue, Qunhua Li. Genome Research 2017. doi: 10.1101/gr.220640.117.
Installation
Download the source package (hicrep_xxx.tar.gz) from Github. Or install it from Bioconductor:
## try http:// if https:// URLs are not supported
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("hicrep")
Rationale of method
This is a 2-step method (Figure1). In Hi-C data it is often difficult to achieve sufficient coverage. When samples are not sufficiently sequenced, the local variation introduced by under-sampling can make it difficult to capture large domain structures. To reduce local variation, we first smooth the contact map before assessing reproducibility. Although a smoothing filter will reduce the individual spatial resolution, it can improve the contiguity of the regions with elevated interaction, consequently enhancing the domain structures. We use a 2D moving window average filter to smooth the Hi-C contact map. This choice is made for the simplicity and fast computation of mean filter, and the rectangular shape of Hi-C compartments.
In the second step, we stratify the Hi-C reads by the distance of contacting loci, calculate the Pearson correlations within each stratum, and then summarize the stratum-specific correlation coefficients into an aggregated statistic. We name it as Stratum-adjusted Correlation Coefficient (SCC). For the methodology details, please refer to our paper on Genome Research.
knitr::opts_knit$set(progress = TRUE, verbose = TRUE) library(hicrep) data("HiCR1") data("HiCR2")
The format of input and Pre-processing
The input are two Hi-C matrices to be compared. The Hi-C matrices should have the dimension $N\times N$.
dim(HiCR1) HiCR1[1:10,1:10]
The function get.scc will first smooth the HiC matrix, with given
neighborhood size parameter $h$, and filter the bins that have
zero counts in both replicates. The arguments includes the two matrices,
the resolution of matrices, smoothing parameter, and the lower bound and
upper bound of interaction distance considered. The resolution is simply
the bin size. Smoothing parameter decides the neighborhood size of
smoothing. Below (Figure 2) is a representation of smoothing neighborhood
for a point $C_{ij}$:
Calculate Stratum-adjusted Correlation Coefficient (SCC)
An example to calculate SCC for a matrix of 1Mb resolution. Smoothing parameter $h$ is set to 2. The lower bound of distance considered is 0 (diagnal), and the upper bound is 5Mb.
scc.out = get.scc(HiCR1, HiCR2, 1000000, 2, 0, 5000000) #SCC score scc.out$scc #Standard deviation of SCC scc.out$std
The output is a list of results including stratum specific Pearson correlations, weight coefficient, SCC and the asymptotic standard deviation of SCC. The last two numbers are the ones we needed in most of the situations.
Smooth the Hi-C matrix with 2D mean filter
The function fast.mean.filter() is a very fast algorithm that applies
2D mean filter to squred matrices such as Hi-C contact mapes. The output
is a smoothed matrix that has the same size with the original matrix.
Here is an example to smooth the matrix with parameter $h = 2$:
smd_mat = fast.mean.filter(HiCR1, 2)
Select the optimal smoothing parameter
To select $h$ objectively, we develop a heuristic procedure to search for the optimal smoothing parameter. Our procedure is designed based on the observation that the correlation between contact maps of replicate samples first increases with the level of smoothness and plateaus when sufficient smoothness is reached.To proceed, we use a pair of reasonably deeply sequenced interaction map as the training data. We randomly sampled 10% of the data, then compute SCC for the sampled dataeach fraction at a series of smoothing parameters in the ascending order. We choose the smallest $h$ at which the increment of the average reproducibility score is less than 0.01. This procedure is repeated ten times, and the mode among the ten $h$’s is picked.
h_hat <- htrain(HiCR1, HiCR2, 1000000, lbr = 0, ubr = 5000000, range = 0:2) h_hat
The above graph shows the change of SCC as the $h$ increases from 0 to 10 for a 40Kb resolution matrix. The parameter $h = 5$ is selected as the optimal smoothing neighborhood size.
Important note: The smoothing parameter selection could be confounded by the sequencing depth. Insufficient sequencing depth data might lead to inflated smoothing neighborhood size. To compare SCC between pairs of replicates that has the same resolution, one shall use the same smoothing parameter.
Train the smoothing parameter could be time-consuming. It is not suggested to train $h$ every time when calculating SCC. For a giving resolution, one could use a deeply sequenced biological replicates to train $h$ (i.e., > 300 million total nubmer of reads for whole chromosome), and use the trained $h$ for other same resolution data. Here we provide some trained $h$ trained based on two replicates of hESC cells from Dixon et al 2015 (GEO accession: GSE52457):
Resolution ($h$): 10K (20), 25K (10), 40k(5), 100k(3), 500k(1 or 2), 1M(0 or 1).
Equalize the total number of reads
In previous section, we mention that sequencing depth could be a confounding
effect. If the total numbers of reads are very different between the two
replicates, it's suggested that one should down-sample the higher sequencing
depth to make it equal to the lower one. The best way to do it is to use the
bam files to do the sub-sampling randomly. In case you only have the matrix
file available, we made a function depth.adj() to do down-sampling from
matrix files.
#check total number of reads before adjustment sum(HiCR1) # sub-sample 200000 total reads DS_HiCR1 <- depth.adj(HiCR1, 200000) #check total number of reads after adjustment sum(DS_HiCR1)
sessionInfo()
#check total number of reads before adjustment sum(HiCR1) # sub-sample 200000 total reads DS_HiCR1 <- depth.adj(HiCR1, 200000) #check total number of reads after adjustment sum(DS_HiCR1)
Computation efficiency
Given a pair of contact maps of human chromosome 1 with bin-size equal to 40kb, it takes 27 seconds on a laptop with 2.6GHz Intel Core i7-6600U and 16Gb of RAM.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
