README.md
In vallotlab/IDclust: Iterative Differential Clustering for single-cell
IDclust
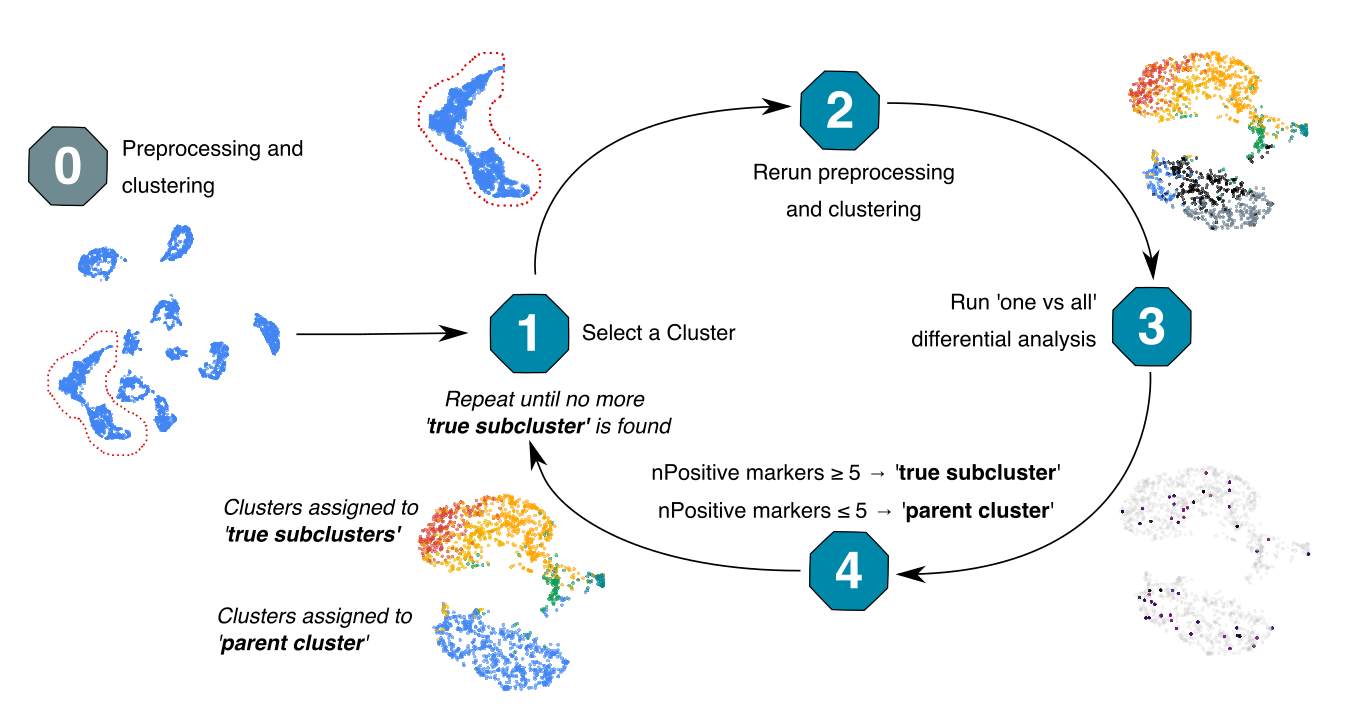
IDclust is an unsupervised method for clustering single-cell datasets (scRNA, scEpigenome). The cells are iteratively re-processed and re-clustered. Subclusters are created provided they are significantly different from each other.
Installation
Install package
if (!require("devtools"))
{
install.packages("devtools", dep=TRUE)
if(!require("devtools")) stop("Package not found")
}
devtools::install_github("vallotlab/IDclust")
For scRNA use
install.packages("Seurat")
For scEpigenomics use
devtools::install_github("vallotlab/ChromSCape")
Usage
Running Iterative Differential Clustering
library(IDclust)
library(Seurat)
data("Seu") # for SingleCellExperiment - data("scExp")
Seu <- iterative_differential_clustering(
Seu, # for SingleCellExperiment - scExp,
output_dir = "./",
plotting = TRUE,
saving = TRUE,
n_dims = 50,
dim_red = "pca",
vizualization_dim_red = "umap",
logFC.th = log2(1.5),
qval.th = 0.01
)
Plotting network
plot_cluster_network(Seu) # for SingleCellExperiment - scExp

vallotlab/IDclust documentation built on July 5, 2024, 3:26 p.m.
IDclust
IDclust is an unsupervised method for clustering single-cell datasets (scRNA, scEpigenome). The cells are iteratively re-processed and re-clustered. Subclusters are created provided they are significantly different from each other.

Installation
Install package
if (!require("devtools"))
{
install.packages("devtools", dep=TRUE)
if(!require("devtools")) stop("Package not found")
}
devtools::install_github("vallotlab/IDclust")
For scRNA use
install.packages("Seurat")
For scEpigenomics use
devtools::install_github("vallotlab/ChromSCape")
Usage
Running Iterative Differential Clustering
library(IDclust)
library(Seurat)
data("Seu") # for SingleCellExperiment - data("scExp")
Seu <- iterative_differential_clustering(
Seu, # for SingleCellExperiment - scExp,
output_dir = "./",
plotting = TRUE,
saving = TRUE,
n_dims = 50,
dim_red = "pca",
vizualization_dim_red = "umap",
logFC.th = log2(1.5),
qval.th = 0.01
)
Plotting network
plot_cluster_network(Seu) # for SingleCellExperiment - scExp

Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
