README.md
In waldronlab/SingleCellMultiModal: Integrating Multi-modal Single Cell Experiment datasets
SingleCellMultiModal
Overview
SingleCellMultiModal is an R package that provides a convenient and
user-friendly representation of multi-modal data using
MultiAssayExperiment. This package introduces a suite of single-cell
multimodal landmark datasets for benchmarking and testing multimodal
analysis methods via the ExperimentHub Bioconductor package. The scope
of this package is to provide efficient access to a selection of
curated, pre-integrated, publicly available landmark datasets for
methods development and benchmarking.
Installation
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("SingleCellMultiModal")
Loading packages
library(SingleCellMultiModal)
library(MultiAssayExperiment)
Citing SingleCellMultiModal
Your citations are crucial in keeping our software free and open source.
To cite our package see the citation (Eckenrode et al. (2023)) in the
Reference section. You may also browse to the publication at PLoS
Computational Biology.
Representation
Users can obtain integrative representations of multiple modalities as a
MultiAssayExperiment, a common core Bioconductor data structure relied
on by dozens of multimodal data analysis packages.
MultiAssayExperiment harmonizes data management of multiple
experimental assays performed on an overlapping set of specimens.
Although originally developed for patient data from multi-omics cancer
studies, the MultiAssayExperiment framework naturally applies also to
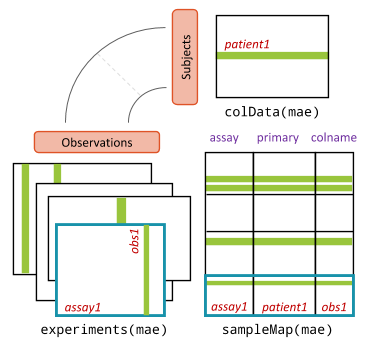
single cells. A schematic of the data structure can be seen below. In
this context, “patients” are replaced by “cells”. We use
MultiAssayExperiment because it provides a familiar user experience by
extending SummarizedExperiment concepts and providing open ended
compatibility with standard data classes present in Bioconductor such as
the SingleCellExperiment.
Contributions
Want to contribute to the SingleCellMultiModal package? We welcome
contributions from the community. Please refer to our Contributing
Guidelines
for more details.
Further resources
For more information on the MultiAssayExperiment data structure,
please refer to Ramos et al. (2017) as well as the MultiAssayExperiment
vignette.
References
Eckenrode, Kelly B, Dario Righelli, Marcel Ramos, Ricard Argelaguet,
Christophe Vanderaa, Ludwig Geistlinger, Aedin C Culhane, et al. 2023.
“Curated Single Cell Multimodal Landmark Datasets for R/Bioconductor.”
*PLoS Comput. Biol.* 19 (8): e1011324.
Ramos, Marcel, Lucas Schiffer, Angela Re, Rimsha Azhar, Azfar Basunia,
Carmen Rodriguez, Tiffany Chan, et al. 2017. “Software for the
Integration of Multiomics Experiments in Bioconductor.” *Cancer Res.* 77
(21): e39–42.
waldronlab/SingleCellMultiModal documentation built on Nov. 3, 2024, 7:32 p.m.
SingleCellMultiModal
Overview
SingleCellMultiModal is an R package that provides a convenient and
user-friendly representation of multi-modal data using
MultiAssayExperiment. This package introduces a suite of single-cell
multimodal landmark datasets for benchmarking and testing multimodal
analysis methods via the ExperimentHub Bioconductor package. The scope
of this package is to provide efficient access to a selection of
curated, pre-integrated, publicly available landmark datasets for
methods development and benchmarking.
Installation
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("SingleCellMultiModal")
Loading packages
library(SingleCellMultiModal)
library(MultiAssayExperiment)
Citing SingleCellMultiModal
Your citations are crucial in keeping our software free and open source. To cite our package see the citation (Eckenrode et al. (2023)) in the Reference section. You may also browse to the publication at PLoS Computational Biology.
Representation
Users can obtain integrative representations of multiple modalities as a
MultiAssayExperiment, a common core Bioconductor data structure relied
on by dozens of multimodal data analysis packages.
MultiAssayExperiment harmonizes data management of multiple
experimental assays performed on an overlapping set of specimens.
Although originally developed for patient data from multi-omics cancer
studies, the MultiAssayExperiment framework naturally applies also to
single cells. A schematic of the data structure can be seen below. In
this context, “patients” are replaced by “cells”. We use
MultiAssayExperiment because it provides a familiar user experience by
extending SummarizedExperiment concepts and providing open ended
compatibility with standard data classes present in Bioconductor such as
the SingleCellExperiment.

Contributions
Want to contribute to the SingleCellMultiModal package? We welcome
contributions from the community. Please refer to our Contributing
Guidelines
for more details.
Further resources
For more information on the MultiAssayExperiment data structure,
please refer to Ramos et al. (2017) as well as the MultiAssayExperiment
vignette.
References
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
