Nothing
README.md
In alevinQC: Generate QC Reports For Alevin Output
alevinQC
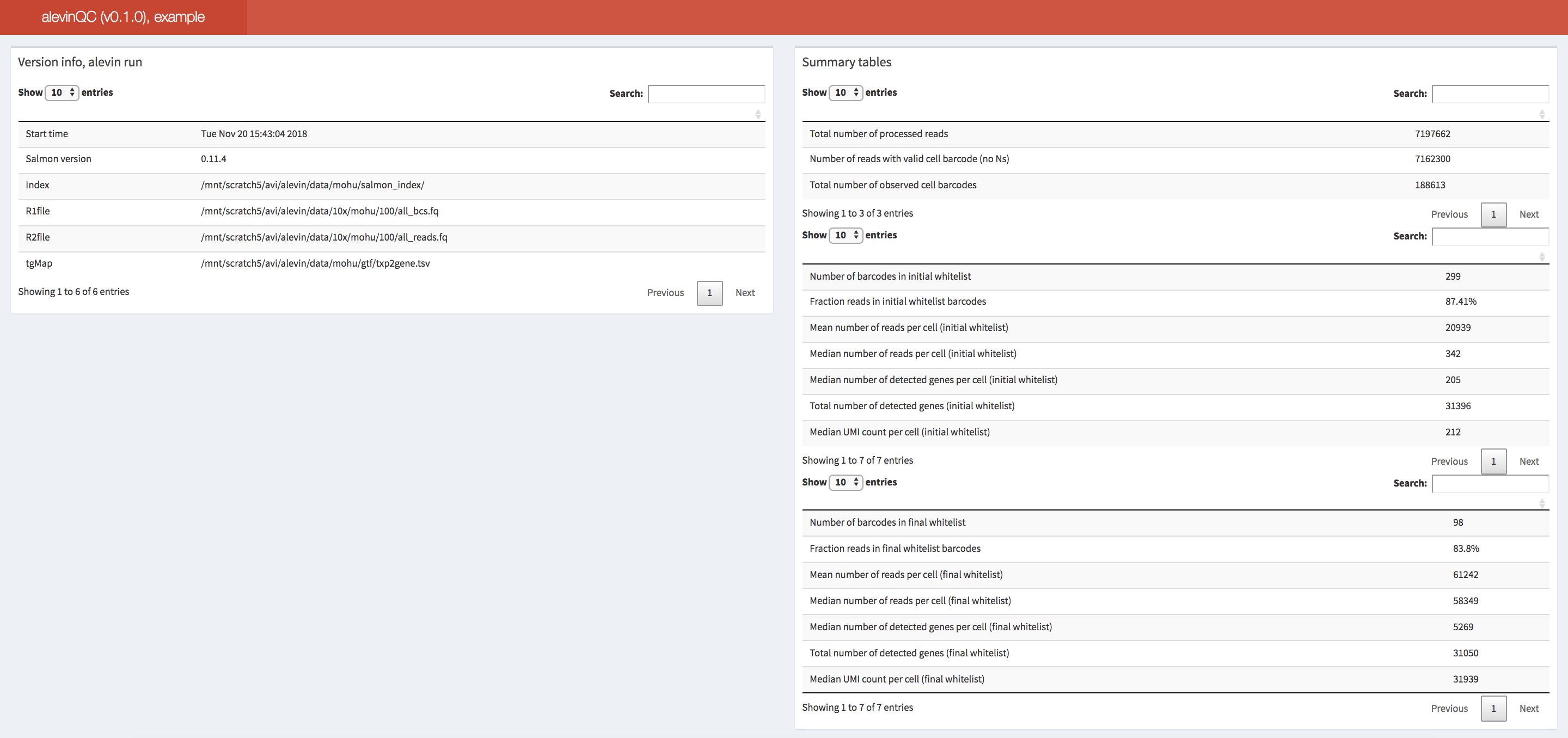
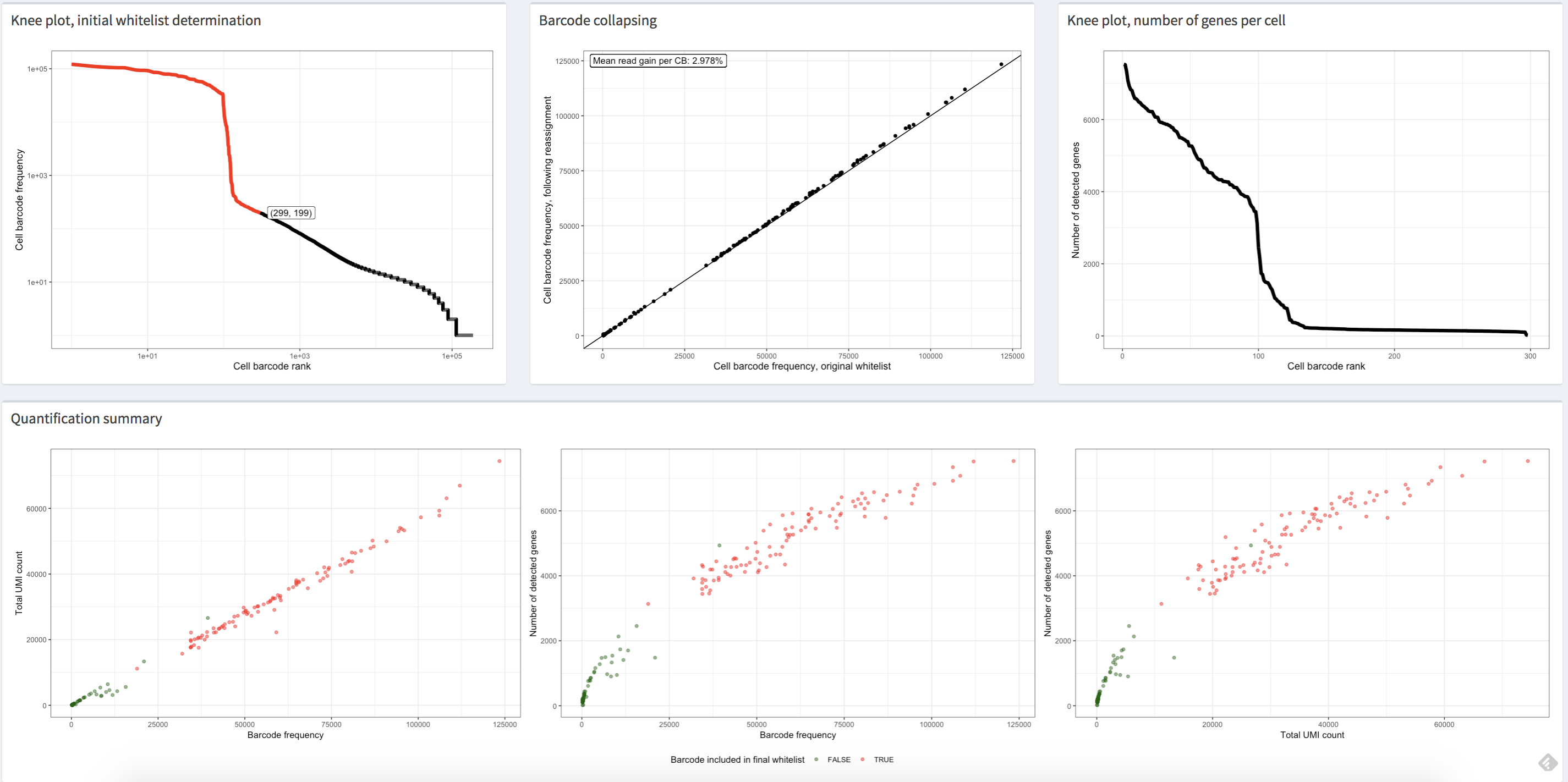
The alevinQC R package provides functionality for generating QC reports
summarizing the output of alevin
(Srivastava et al., Genome Biology 20:65 (2019)). The reports can
be generated in html or pdf format, or as R/Shiny applications.
Installation:
alevinQC is available from
Bioconductor, and can be
installed using the BiocManager CRAN package:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("alevinQC")
Note that alevinQC v 1.1 or newer is required to process output from Salmon version 0.14.0 or newer.
Example usage:
alevinQCReport(baseDir = system.file("extdata/alevin_example_v0.14",
package = "alevinQC"),
sampleId = "testSample",
outputFile = "alevinReport.html",
outputFormat = "html_document",
outputDir = tempdir(), forceOverwrite = TRUE)
For more information, we refer to the package vignette.
Try the alevinQC package in your browser
Any scripts or data that you put into this service are public.
alevinQC documentation built on Feb. 4, 2021, 2:01 a.m.
alevinQC
The alevinQC R package provides functionality for generating QC reports
summarizing the output of alevin
(Srivastava et al., Genome Biology 20:65 (2019)). The reports can
be generated in html or pdf format, or as R/Shiny applications.
Installation:
alevinQC is available from
Bioconductor, and can be
installed using the BiocManager CRAN package:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("alevinQC")
Note that alevinQC v 1.1 or newer is required to process output from Salmon version 0.14.0 or newer.
Example usage:
alevinQCReport(baseDir = system.file("extdata/alevin_example_v0.14",
package = "alevinQC"),
sampleId = "testSample",
outputFile = "alevinReport.html",
outputFormat = "html_document",
outputDir = tempdir(), forceOverwrite = TRUE)
For more information, we refer to the package vignette.


Try the alevinQC package in your browser
Any scripts or data that you put into this service are public.
Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
