README.md
In shangll123/SpatialPCA: Spatially Aware Dimension Reduction for Spatial Transcriptomics
SpatialPCA
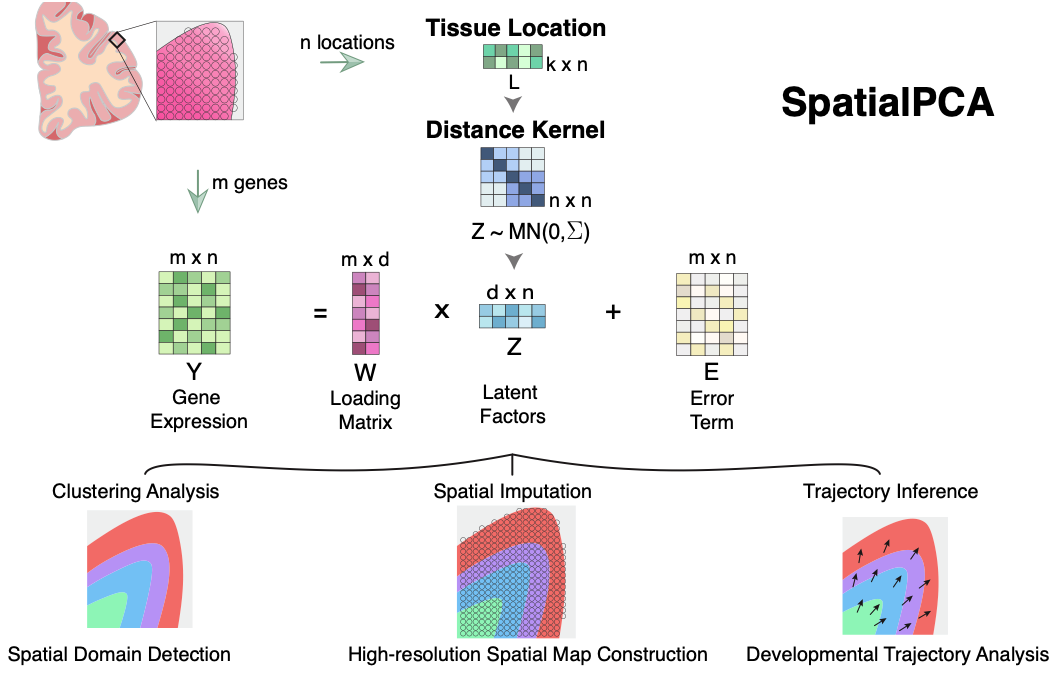
SpatialPCA is a spatially aware dimension reduction method that aims to infer a low dimensional representation of the gene expression data in spatial transcriptomics. SpatialPCA builds upon the probabilistic version of PCA, incorporates localization information as additional input, and uses a kernel matrix to explicitly model the spatial correlation structure across tissue locations. SpatialPCA is implemented as an open-source R package, freely available at www.xzlab.org/software.html.
Install the Package
You can install the current version of SpatialPCA from GitHub with:
library(devtools)
install_github("shangll123/SpatialPCA")
Package Tutorial
Please see the SpatialPCA tutorial website.
The tutorial includes main example codes for multiple spatial transcriptomics datasets (e.g. DLPFC, Slide-Seq cerebellum, Slide-Seq V2 hippocampus, Human breast tumor, and Vizgen MERFISH.)
Other analysis codes for this project can be found here.
Example data can be found here: https://drive.google.com/drive/folders/1Ibz5uNsFKHJ4roPpaec5nPL_EBF3-wxY?usp=share_link.
Operating systems (version 1.3.0 SpatialPCA) tested on:
macOS Catalina 10.15.7
Ubuntu 18.04.5 LTS (Bionic Beaver)
CentOS Linux 7 (Core)
License
SpatialPCA is licensed under the GNU General Public License v3.0.
Citation
Lulu Shang, and Xiang Zhou (2022). Spatially aware dimension reduction for spatial transcriptomics. Nature Communications.
doi: https://www.nature.com/articles/s41467-022-34879-1
shangll123/SpatialPCA documentation built on April 17, 2024, 3:15 a.m.
SpatialPCA
SpatialPCA is a spatially aware dimension reduction method that aims to infer a low dimensional representation of the gene expression data in spatial transcriptomics. SpatialPCA builds upon the probabilistic version of PCA, incorporates localization information as additional input, and uses a kernel matrix to explicitly model the spatial correlation structure across tissue locations. SpatialPCA is implemented as an open-source R package, freely available at www.xzlab.org/software.html.

Install the Package
You can install the current version of SpatialPCA from GitHub with:
library(devtools)
install_github("shangll123/SpatialPCA")
Package Tutorial
Please see the SpatialPCA tutorial website.
The tutorial includes main example codes for multiple spatial transcriptomics datasets (e.g. DLPFC, Slide-Seq cerebellum, Slide-Seq V2 hippocampus, Human breast tumor, and Vizgen MERFISH.)
Other analysis codes for this project can be found here. Example data can be found here: https://drive.google.com/drive/folders/1Ibz5uNsFKHJ4roPpaec5nPL_EBF3-wxY?usp=share_link.
Operating systems (version 1.3.0 SpatialPCA) tested on:
macOS Catalina 10.15.7
Ubuntu 18.04.5 LTS (Bionic Beaver)
CentOS Linux 7 (Core)
License
SpatialPCA is licensed under the GNU General Public License v3.0.
Citation
Lulu Shang, and Xiang Zhou (2022). Spatially aware dimension reduction for spatial transcriptomics. Nature Communications.
doi: https://www.nature.com/articles/s41467-022-34879-1
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
