README.md
In tkonopka/ExpCube: Analysis of cell/stimulus/gene expression data
ExpCube
ExpCube is an R package for analysis of gene expression data, e.g. RNA-seq data, from experiments with large numbers of samples and conditions.
The package is developed to support analysis in Gapp et al
Overview
Gene expression analysis involves quantifying mRNA abundance and calling differential expression. A simple experimental design consists of measurements in two groups, e.g. replicates of treated and untreated samples. Such data can be arranged in a 2d matrix with axes denoting genes and samples.
There exist several approaches to compare expression profiles between pairs of sample groups.
More complex experimental designs might involve several related treatment conditions or cell types. For example, experiments might investigate various drug doses, time profiles, or environmental stimulation. In this package, these will be referred to as 'stimuli'. This term is meant to encompass all types of environmental factors and/or treatments.
In principle, it is possible to force data from a complex experiment into a 2d matrix, as above. Indeed, from a data storage perspective, this is the natural structure for all expression data. However, the flat structure hides dependence between samples. A different approach is thus useful. This package relies on a 3d organization structure - a cube with axes representing genes, cell types, and stimuli.
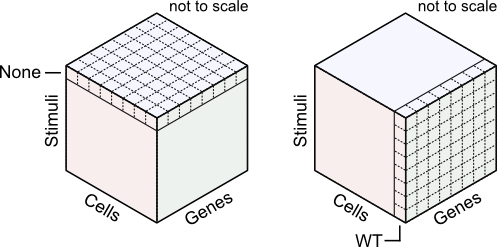
From this viewpoint, 'simple' experimental designs are slices in the 3d expression cube. But the 3d structure can help visualize more complex experiments as well. For example, comparisons of two cell types (mutant vs. wild type) in response to a stimulus (treated vs. untreated) can be displayed as four blocks.
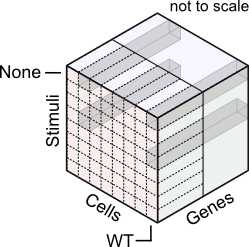
Such analysis might rely on the whole gene profile, or might emphasize subset of genes.
Package
The package ExpCube is built around the 3d cube organization of expression data. The sections below contain a brief outline of the organization of the package functions.
Data
The package data-manipulation functions operate on objects that hold "gene"-expression data. ("Gene" here can refer to any genomic region of interest).
In most cases, the primary data objects are lists with three data frames. Each data frame contains a column with gene name and several columns with sample names. One of the three data frames contains 'canonical' gene expression estimates. The other two data frames hold low- and high- estimates of gene expression. Together, these objects encode expression 'confidence' intervals. (The precise interpretation of the intervals is analysis-dependent and is determined by the user of the package.)
In some cases, package functions operate on simple expression matrices.
Annotations
Several package functions require one or more sample-annotation objects.
A common required object is a definition of sample groups. This should be a named list of character vectors. Each character vector should contain samples (e.g. replicates) that belong to a group.
Other common required objects are definitions of reference samples. These should be character vectors with names. Names of the vector elements should identify samples. Values should identify sample groups. For example, a vector might link a mutant sample stimulated with agent A to a group containing the same mutant cell line with mock stimulation. As another example, another vector might link the same mutant sample stimulated with agent A with a wild type sample stimulated with the same agent.
Visualizations
The package contains some customized plot functions (box plots, scatter plots, heatmaps, etc.). These functions provide styling through Rcssplot.
Vignettes
The package includes vignettes showcasing the package capabilities. These examples are limited, however, by the size of dataset that can be reasonably attached to a package.
For a fuller view of the package capabilities, please refer to the scripts associated with the publication.
Development
The package code is available under a GPL-2 license.
The package depends on some third-party packages.
- data.table - manipulation of data frames.
- sm - smoothing (used in visualization)
- DNAcopy - segmentation of signals
- Rcssplot - customization of graphics functions.
To cite the package codebase or data organization principles, please link to Gapp et al.
tkonopka/ExpCube documentation built on May 31, 2019, 3:44 p.m.
ExpCube
ExpCube is an R package for analysis of gene expression data, e.g. RNA-seq data, from experiments with large numbers of samples and conditions.
The package is developed to support analysis in Gapp et al
Overview
Gene expression analysis involves quantifying mRNA abundance and calling differential expression. A simple experimental design consists of measurements in two groups, e.g. replicates of treated and untreated samples. Such data can be arranged in a 2d matrix with axes denoting genes and samples.

There exist several approaches to compare expression profiles between pairs of sample groups.
More complex experimental designs might involve several related treatment conditions or cell types. For example, experiments might investigate various drug doses, time profiles, or environmental stimulation. In this package, these will be referred to as 'stimuli'. This term is meant to encompass all types of environmental factors and/or treatments.
In principle, it is possible to force data from a complex experiment into a 2d matrix, as above. Indeed, from a data storage perspective, this is the natural structure for all expression data. However, the flat structure hides dependence between samples. A different approach is thus useful. This package relies on a 3d organization structure - a cube with axes representing genes, cell types, and stimuli.

From this viewpoint, 'simple' experimental designs are slices in the 3d expression cube. But the 3d structure can help visualize more complex experiments as well. For example, comparisons of two cell types (mutant vs. wild type) in response to a stimulus (treated vs. untreated) can be displayed as four blocks.

Such analysis might rely on the whole gene profile, or might emphasize subset of genes.
Package
The package ExpCube is built around the 3d cube organization of expression data. The sections below contain a brief outline of the organization of the package functions.
Data
The package data-manipulation functions operate on objects that hold "gene"-expression data. ("Gene" here can refer to any genomic region of interest).
In most cases, the primary data objects are lists with three data frames. Each data frame contains a column with gene name and several columns with sample names. One of the three data frames contains 'canonical' gene expression estimates. The other two data frames hold low- and high- estimates of gene expression. Together, these objects encode expression 'confidence' intervals. (The precise interpretation of the intervals is analysis-dependent and is determined by the user of the package.)
In some cases, package functions operate on simple expression matrices.
Annotations
Several package functions require one or more sample-annotation objects.
A common required object is a definition of sample groups. This should be a named list of character vectors. Each character vector should contain samples (e.g. replicates) that belong to a group.
Other common required objects are definitions of reference samples. These should be character vectors with names. Names of the vector elements should identify samples. Values should identify sample groups. For example, a vector might link a mutant sample stimulated with agent A to a group containing the same mutant cell line with mock stimulation. As another example, another vector might link the same mutant sample stimulated with agent A with a wild type sample stimulated with the same agent.
Visualizations
The package contains some customized plot functions (box plots, scatter plots, heatmaps, etc.). These functions provide styling through Rcssplot.
Vignettes
The package includes vignettes showcasing the package capabilities. These examples are limited, however, by the size of dataset that can be reasonably attached to a package.
For a fuller view of the package capabilities, please refer to the scripts associated with the publication.
Development
The package code is available under a GPL-2 license.
The package depends on some third-party packages.
- data.table - manipulation of data frames.
- sm - smoothing (used in visualization)
- DNAcopy - segmentation of signals
- Rcssplot - customization of graphics functions.
To cite the package codebase or data organization principles, please link to Gapp et al.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
