Nothing
inst/shinyapp/helper/data_origin.md
In UCSCXenaShiny: Interactive Analysis of UCSC Xena Data
- In this step, you can check which UCSC Xena hub and dataset each type of Molecular profile comes from for downstream analysis. You can click on the provided hub and dataset links at the bottom to learn more details about them;
- For some data types, there are multiple datasets available based on factors such as standardization methods, sequencing platforms, and others;
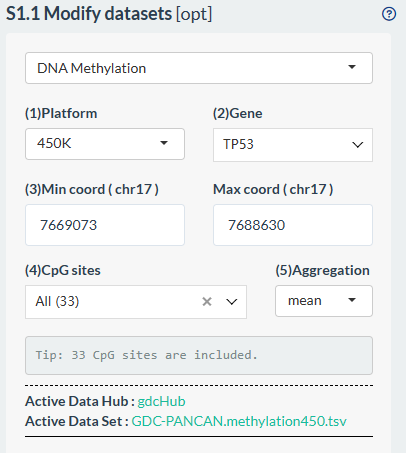
- Noteworthily, we provide more personlized options for DNA methylation type (Only TCGA);
- Select one methylation platform;
- Select one gene;
- Select CpG sites through chromosomal coordinates. By default, it is the range of all sites for that gene;
- Further manually select CpG sites based on above coordinates. By default, all sites will be selected
- Select the aggregation method for calculating gene-level methylation. By default, the mean of the selected CpG sites will be calculated.
- See more details in UCSCXenaShiny v2 book.
Try the UCSCXenaShiny package in your browser
Any scripts or data that you put into this service are public.
UCSCXenaShiny documentation built on Aug. 8, 2025, 7:19 p.m.
- In this step, you can check which UCSC Xena hub and dataset each type of Molecular profile comes from for downstream analysis. You can click on the provided hub and dataset links at the bottom to learn more details about them;
- For some data types, there are multiple datasets available based on factors such as standardization methods, sequencing platforms, and others;
- Noteworthily, we provide more personlized options for DNA methylation type (Only TCGA);
- Select one methylation platform;
- Select one gene;
- Select CpG sites through chromosomal coordinates. By default, it is the range of all sites for that gene;
- Further manually select CpG sites based on above coordinates. By default, all sites will be selected
- Select the aggregation method for calculating gene-level methylation. By default, the mean of the selected CpG sites will be calculated.

- See more details in UCSCXenaShiny v2 book.
Try the UCSCXenaShiny package in your browser
Any scripts or data that you put into this service are public.
Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
